




|
 
|

|
Here on this This Week's Finds I've been talking about the future and what it might hold. But any vision of the future that ignores biotechnology is radically incomplete. Just look at this week's news! They've 'hacked the genome':
Or maybe they've 'hijacked the genetic code':
What exactly have they done? These articles explain it quite well... but it's so cool I can't resist talking about it.
Basically, some scientists from Harvard and MIT have figured out how to go through the whole genome of a bacterium and change every occurrence of one codon to some other codon. It's a bit like the 'global search and replace' feature of a word processor. You know: that trick where you can take a document and replace one word with another every place it appears.
To understand this better, it helps to know a tiny bit about the genetic code. You may know this stuff, but let's quickly review.
DNA is a double-stranded helix bridged by pairs of bases, which come in 4 kinds:
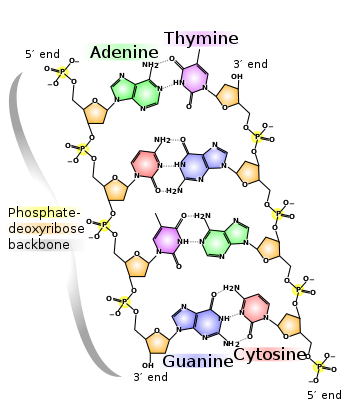
Because of how they're shaped, A can only connect to T:

while C can only connect to G:

So, all the information in the DNA is contained in the list of bases down either side of the helix. You can think of it as a long string of 'letters', like this:
ATCATTCAGCTTATGC...
This long string consists of many sections, which are the instructions to make different proteins. In the first step of the protein manufacture process, a section of this string copied to a molecule called 'messenger RNA'. In this stage, the T gets copied to uracil, or U. The other three base pairs stay the same.
Here's some messenger RNA:
You'll note that the bases come in groups of three. Each group is called a 'codon', because it serves as the code for a specific amino acid. A protein is built as a string of amino acids, which then curls up into a complicated shape.
Here's how the genetic code works:
The three-letter names like Phe and Leu are abbreviations for amino acids: phenylalanine, leucine and so on.
While there are 43 = 64 codons, they code for only 20 amino acids. So, typically more than one codon codes for the same amino acid. If you look at the chart, you'll see one exception is methionine, which is encoded only by AUG. AUG is also the 'start codon', which tells the cell where a protein starts. So, methionine shows up at the start of every protein, at least at first. It's usually removed later in the protein manufacture process.
There are also three 'stop codons', which mark the end of a protein. They have cute names:
UAG was named after Harris Bernstein, whose last name means 'amber' in German. The other two names were just a way of continuing the joke.
And now we're ready to understand how a team of scientists led by Farren J. Isaacs and George M. Church are 'hacking the genome'. They're going through the DNA of the common E. coli bacterium and replacing every instance of amber with opal!
This is a lot more work than the word processor analogy suggests. They need to break the DNA into lots of fragments, change amber to opal in these fragments, and put them back together again. Read Ed Young's article for more.
So, they're not actually done yet.
But when they're done, they'll have an E. coli bacterium with no amber codons, just opal. But it'll act just the same as ever, since amber and opal are both stop codons.
That's a lot of work for no visible effect. What's the point?
The point is that they'll have freed up the codon amber for other purposes! This will let them do various further tricks.
First, with some work, they could make amber code for a new, unnatural amino acid that's not one of the usual 20. This sounds like a lot of work, since it requires tinkering with the cell's mechanisms for translating codons into amino acids: specifically, its set of transfer RNA and synthetase molecules. But this has already been done! Back in 1990, Jennifer Normanly found a viable mutant strain of E. coli that 'reads through' the amber codon, not stopping the protein there as it should. People have taken advantage of this to create E. coli where amber codes for a new amino acid:
But I guess getting an E. coli that's completely free of amber codons would let us put amber codons only where we want them, getting better control of the situation.
Second, tweaking the genetic code this way could yield a strain of E. coli that's unable to 'breed' with the normal kind. This could increase the safety of genetic engineering. Of course bacteria are asexual, so they don't precisely 'breed'. But they do something similar: they exchange genes with each other! Three of the most popular ways are:
Thanks to these tricks, drug resistance and other traits can hop from one species of bug to another. So, for the sake of safe experiments, it would be nice to have a strain of bacteria whose genetic code was so different from others that it couldn't share DNA.
And third, a bacterium with a modified genetic code could be resistant to viruses! I hadn't known it, but the biotech firm Genzyme was shut down for three months and lost millions of dollars when its bacteria were hit by a virus.
This third application reminds me of a really spooky story by Greg Egan, called "The Moat". In it, a detective discovers evidence that some people have managed to alter their genetic code. The big worry is that they could then set loose a virus that would kill everyone in the world except them.
That's a scary idea, and one that just became a bit more practical... though so far only for E. coli, not H. sapiens.
So, I've got some questions for the biologists out there.
A virus that attacks bacteria is called a bacteriophage---or affectionately, a 'phage'. Here's a picture of one:
Isn't it cute?
Whoops—that wasn't one of the questions. Here are my questions for biologists:
and more generally:
Also, on a more technical note:
Finally, I can't resist mentioning something amazing I just read. I said that our body uses 20 amino acids, and that 'opal' serves a stop codon. But neither of these are the whole truth! Sometimes opal codes for a 21st amino acid, called selenocysteine. And this one is different from the rest. Most amino acids contain carbon, hydrogen, oxygen and nitrogen, and cysteine contains sulfur, but selenocysteine contains... you guessed it... selenium!
Selenium is right below sulfur on the periodic table, so it's sort of similar. If you eat too much selenium, your breath starts smelling like garlic and your hair falls out. Horses have died from the stuff. But it's also an essential trace element: you have about 15 milligrams in your body. We use it in various proteins, which are called.... you guessed it... selenoproteins!
So, a few more questions:
Finally, here's the new paper that all the fuss is about. It's not free, but you can read the abstract for free:
For more discussion go to my blog, Azimuth.
Pessimists should be reminded that part of their pessimism is an inability to imagine the creative ideas of the future - Brian Eno
© 2011 John Baez
baez@math.removethis.ucr.andthis.edu

|
 
|

|