*Guest post by [Pablo Andres-Martinez](https://www.inf.ed.ac.uk/people/students/Pablo_Andres_Martinez.html) and [Sophie Raynor](https://www.hoppinger.com/)*
In the final instalment of the [Applied Category Theory
seminar](http://www.appliedcategorytheory.org/school/), we discussed the
2014 paper [*"Theory-independent limits on correlations from generalized
Bayesian
networks"*](http://iopscience.iop.org/article/10.1088/1367-2630/16/11/113043/meta)
by Henson, Lal and Pusey.
In this post, we'll give a short introduction to Bayesian networks,
explain why quantum mechanics means that one may want to generalise
them, and present the main results of the paper. That's a lot to cover,
and there won't be a huge amount of category theory, but we hope to give
the reader some intuition about the issues involved, and
[another](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
example of monoidal categories used in causal theory.
## **Introduction**
Bayesian networks are a graphical modelling tool used to show how random
variables interact. A Bayesian network consists of a pair $(G,P)$ of
directed acyclic graph (DAG) $G$ together with a joint probability
distribution $P$ on its nodes, satisfying the [*Markov
condition*](#markov-condition). Intuitively the graph describes a flow of
information.
The Markov condition says that the system doesn't have *memory*. That
is, the distribution on a given node $Y$ is only dependent on the
distributions on the nodes $X$ for which there is an edge
$X \rightarrow Y$. Consider the following chain of binary events. In
spring, the pollen in the air may cause someone to have an allergic
reaction that may make them sneeze.
> 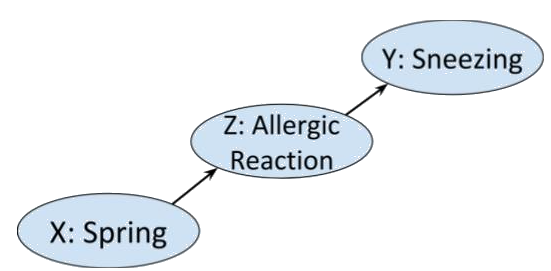 In this case the Markov condition says that given that you know that
someone is having an allergic reaction, whether or not it is spring is
not going to influence your belief about the likelihood of them
sneezing. Which seems sensible.
Bayesian networks are useful
- as an inference tool, thanks to [belief
propagation](https://arxiv.org/abs/1201.4724) algorithms,
- and because, given a Bayesian network $(G,P)$, we can describe ['*d-separation*'](#d-separation) properties on $G$ which enable us to discover conditional
independences in $P$.
It is this second point that we'll be interested in here.
Before getting into the details of the paper, let's try to motivate this
discussion by explaining its title: *"Theory-independent limits on
correlations from generalized Bayesian networks\"* and giving a little
more background to the problem it aims to solve.
Crudely put, the paper aims to generalise a method that assumes
*classical mechanics* to one that holds in *quantum* and more general
theories.
Classical mechanics rests on two intuitively reasonable and desirable
assumptions, together called *local causality*,
- **Causality:**
Causality is usually treated as a physical primitive.
Simply put it is the principle that there is a (partial) ordering
of events in space time. In order to have information flow from
event $A$ to event $B$, $A$ *must* be in the past of $B$.
Physicists often define causality in terms of a *discarding*
principle: If we ignore the outcome of a physical process, it
doesn't matter what process has occurred. Or, put another way, the
outcome of a physical process doesn't change the initial conditions.
- **Locality:**
Locality is the assumption that, at any given instant, the values of
any particle's properties are independent of any other particle.
Intuitively, it says that particles are individual entities that can
be understood in isolation of any other particle.
Physicists usually picture particles as having a private list of
*numbers* determining their properties. The principle of locality
would be violated if any of the entries of such a list were *a
function* whose domain is another particle's property values.
In 1935 [Einstein, Podolsky and
Rosen](https://journals.aps.org/pr/pdf/10.1103/PhysRev.47.777) showed
that quantum mechanics (which was a recently born theory) predicted that
a pair of particles could be prepared so that applying an action on one
of them would instantaneously affect the other, no matter how distant in
space they were, thus contradicting local causality. This seemed so
unreasonable that the authors presented it as evidence that quantum
mechanics was wrong.
But Einstein was wrong. In 1964, [John S.
Bell](http://elib.bsu.by/bitstream/123456789/154530/1/1964-001%20P%20Bell%20-%20On%20the%20Einstein%20Podolsky%20Rosen%20paradox.pdf) set the bases for an
experimental test that would demonstrate that Einstein's "spooky action
at a distance" (Einstein's own words), now known as *entanglement*, was
indeed real. Bell's experiment has been replicated countless of times
and has plenty of variations. [This
video](https://www.youtube.com/watch?v=MUrXhAxGOWU) gives a detailed
explanation of one of these experiments, for a non-physicist audience.
But then, if acting on a particle has an instantaneous effect on a
distant point in space, one of the two principle above is violated: On
one hand, if we acted on both particles at the same time, each action
being a distinct event, both would be affecting each other's result, so
it would not be possible to decide on an ordering; causality would be
broken. The other option would be to reject locality: a property's value
may be given by *a function*, so the resulting value may
*instantaneously* change when the distant 'domain' particle is altered.
In that case, the particles' information was never separated in space,
as they were never truly isolated, so causality is preserved.
Since causality is integral to our understanding of the world and forms
the basis of scientific reasoning, the standard interpretation of
quantum mechanics is to accept non-locality.
The definition of Bayesian networks implies a [discarding
principle](#operational-probabilistic-theories) and hence there is a formal sense in which they are
causal (even if, as we shall see, the correlations they model do not
always reflect the temporal order). Under this interpretation, the
causal theory Bayesian networks describe is classical. Precisely, they
can only model probability distributions that satisfy local causality.
Hence, in particular, they are not sufficient to model all physical
correlations.
The goal of the paper is to develop a framework that generalises Bayesian
networks and d-separation results, so that we can still use graph
properties to reason about conditional dependence under any
given causal theory, be it classical, quantum, or even more general. In
particular, this theory will be able to handle all physically observed
correlations, and all theoretically postulated correlations.
Though category theory is not mentioned explicitly, the authors achieve
their goal by using the categorical framework of [*operational
probablistic theories* (OPTs)](https://arxiv.org/pdf/0908.1583.pdf).
## **Bayesian networks and d-separation**
Consider the situation in which we have three Boolean random variables.
Alice is either *sneezing* or she is not, she either has a a *fever* or
she does not, and she may or may not have *flu*.
Now, flu can cause both sneezing and fever, that is
$$P(sneezing \ | \ flu ) \neq P( sneezing) \ \text{ and likewise } \ P(fever \ | \ flu ) \neq P( fever)$$
so we could represent this graphically as
>
In this case the Markov condition says that given that you know that
someone is having an allergic reaction, whether or not it is spring is
not going to influence your belief about the likelihood of them
sneezing. Which seems sensible.
Bayesian networks are useful
- as an inference tool, thanks to [belief
propagation](https://arxiv.org/abs/1201.4724) algorithms,
- and because, given a Bayesian network $(G,P)$, we can describe ['*d-separation*'](#d-separation) properties on $G$ which enable us to discover conditional
independences in $P$.
It is this second point that we'll be interested in here.
Before getting into the details of the paper, let's try to motivate this
discussion by explaining its title: *"Theory-independent limits on
correlations from generalized Bayesian networks\"* and giving a little
more background to the problem it aims to solve.
Crudely put, the paper aims to generalise a method that assumes
*classical mechanics* to one that holds in *quantum* and more general
theories.
Classical mechanics rests on two intuitively reasonable and desirable
assumptions, together called *local causality*,
- **Causality:**
Causality is usually treated as a physical primitive.
Simply put it is the principle that there is a (partial) ordering
of events in space time. In order to have information flow from
event $A$ to event $B$, $A$ *must* be in the past of $B$.
Physicists often define causality in terms of a *discarding*
principle: If we ignore the outcome of a physical process, it
doesn't matter what process has occurred. Or, put another way, the
outcome of a physical process doesn't change the initial conditions.
- **Locality:**
Locality is the assumption that, at any given instant, the values of
any particle's properties are independent of any other particle.
Intuitively, it says that particles are individual entities that can
be understood in isolation of any other particle.
Physicists usually picture particles as having a private list of
*numbers* determining their properties. The principle of locality
would be violated if any of the entries of such a list were *a
function* whose domain is another particle's property values.
In 1935 [Einstein, Podolsky and
Rosen](https://journals.aps.org/pr/pdf/10.1103/PhysRev.47.777) showed
that quantum mechanics (which was a recently born theory) predicted that
a pair of particles could be prepared so that applying an action on one
of them would instantaneously affect the other, no matter how distant in
space they were, thus contradicting local causality. This seemed so
unreasonable that the authors presented it as evidence that quantum
mechanics was wrong.
But Einstein was wrong. In 1964, [John S.
Bell](http://elib.bsu.by/bitstream/123456789/154530/1/1964-001%20P%20Bell%20-%20On%20the%20Einstein%20Podolsky%20Rosen%20paradox.pdf) set the bases for an
experimental test that would demonstrate that Einstein's "spooky action
at a distance" (Einstein's own words), now known as *entanglement*, was
indeed real. Bell's experiment has been replicated countless of times
and has plenty of variations. [This
video](https://www.youtube.com/watch?v=MUrXhAxGOWU) gives a detailed
explanation of one of these experiments, for a non-physicist audience.
But then, if acting on a particle has an instantaneous effect on a
distant point in space, one of the two principle above is violated: On
one hand, if we acted on both particles at the same time, each action
being a distinct event, both would be affecting each other's result, so
it would not be possible to decide on an ordering; causality would be
broken. The other option would be to reject locality: a property's value
may be given by *a function*, so the resulting value may
*instantaneously* change when the distant 'domain' particle is altered.
In that case, the particles' information was never separated in space,
as they were never truly isolated, so causality is preserved.
Since causality is integral to our understanding of the world and forms
the basis of scientific reasoning, the standard interpretation of
quantum mechanics is to accept non-locality.
The definition of Bayesian networks implies a [discarding
principle](#operational-probabilistic-theories) and hence there is a formal sense in which they are
causal (even if, as we shall see, the correlations they model do not
always reflect the temporal order). Under this interpretation, the
causal theory Bayesian networks describe is classical. Precisely, they
can only model probability distributions that satisfy local causality.
Hence, in particular, they are not sufficient to model all physical
correlations.
The goal of the paper is to develop a framework that generalises Bayesian
networks and d-separation results, so that we can still use graph
properties to reason about conditional dependence under any
given causal theory, be it classical, quantum, or even more general. In
particular, this theory will be able to handle all physically observed
correlations, and all theoretically postulated correlations.
Though category theory is not mentioned explicitly, the authors achieve
their goal by using the categorical framework of [*operational
probablistic theories* (OPTs)](https://arxiv.org/pdf/0908.1583.pdf).
## **Bayesian networks and d-separation**
Consider the situation in which we have three Boolean random variables.
Alice is either *sneezing* or she is not, she either has a a *fever* or
she does not, and she may or may not have *flu*.
Now, flu can cause both sneezing and fever, that is
$$P(sneezing \ | \ flu ) \neq P( sneezing) \ \text{ and likewise } \ P(fever \ | \ flu ) \neq P( fever)$$
so we could represent this graphically as
> 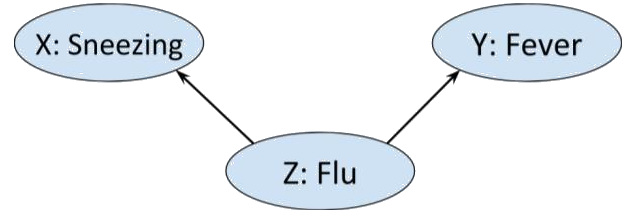 Moreover, intuitively we wouldn't expect there to be any other edges in
the above graph. Sneezing and fever, though correlated - each is more
likely if Alice has flu - are not direct causes of each other. That is,
$$P(sneezing \ | \ fever ) \neq P(sneezing) \ \text{ but } \ P(sneezing \ | \ fever, \ flu ) = P(sneezing \ | \ flu).$$
### *Bayesian networks*
Let $G$ be a *directed acyclic graph* or *DAG* $G$. (Here a directed
graph is a [presheaf on
($\bullet \rightrightarrows \bullet$)](https://golem.ph.utexas.edu/category/2008/01/mark_weber_on_nerves_of_catego.html)).
The set $Pa(Y)$ of *parents* of a node $Y$ of $G$ contains those nodes
$X$ of $G$ such that there is a directed edge $X \to Y$.
So, in the example above $Pa(flu) = \emptyset$ while
$Pa(fever) = Pa(sneezing) = \{ flu \}$.
To each node $X$ of a directed graph $G$, we may associate a random
variable, also denoted $X$. If $V$ is the set of nodes of $G$ and
$(x_X)_{X \in V}$ is a choice of value $x_X$ for each node $X$, such
that $y$ is the chosen value for $Y$, then $pa(y)$ will denote the
$Pa(Y)$-tuple of values $(x_X)_{X \in Pa(Y)}$.
To define Bayesian networks, and establish the notation, let's revise
some probability basics.
Let $P(x,y \ | \ z)$ mean $P(X = x \text{ and } \ Y = y \ | \ Z = z)$,
the *probability that $X$ has the value $x$, and $Y$ has the value $y$
given that $Z$ has the value $z$*. Recall that this is given by
$$P(x,y \ |\ z) = \frac{ P(x,y,z) }{P(z)}.$$
The *chain rule* says that, given a value $x$ of $X$ and sets of values
$\Omega, \Lambda$ of other random variables,
$$P(x, \Omega \ | \ \Lambda) = P( x \ | \ \Lambda) P( \Omega \ | \ x, \Lambda).$$
Random variables $X$ and $Y$ are said to be *conditionally independent
given $Z$*, written $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$, if for all values
$x$ of $X$, $y$ of $Y$ and $z$ of $Z$
$$P(x,y \ | \ z) = P(x \ | \ z) P(y \ | \ z).$$
By the chain rule this is equivalent to
$$P(x \ | \ y,z ) = P (x \ | \ z) , \ \forall x,y, z.$$
More generally, we may replace $X,Y$ and $Z$ with sets of random
variables. So, in the special case that $Z$ is empty, then $X$ and $Y$
are independent if and only if $P(x, y) = P(x)P(y)$ for all $x,y$.
#### Markov condition
A joint probability distribution $P$ on the nodes of a DAG $G$ is said
to satisfy the Markov condition if for any set of random
variable $\{X_i\}_{i = 1}^n$ on the nodes of $G$, with choice of values
$\{x_i\}_{i = 1}^n$
$$P(x_i, \dots, x_n) = \prod_{i = 1}^n P(x_i \ | \ {pa(x_i)}).$$
So, for the flu, fever and sneezing example above, a distribution $P$
satisfies the Markov condition if
$$P(flu, \ fever, \ sneezing) = P(fever \ | \ flu) P(sneezing \ | \ flu) P(flu).$$
A Bayesian network is defined as a pair $(G,P)$ of a DAG $G$ and a joint
probability distribution $P$ on the nodes of $G$ that satisfies the
Markov condition with respect to $G$. This means that each node in a
Bayesian network is conditionally independent, given its parents, of any
of the remaining nodes.
In particular, given a Bayesian network $(G,P)$ such that there is a
directed edge $X \to Y$, the Markov condition implies that
$$\sum_{y} P(x,y) = \sum_y P(x) P(y \ | \ x) = P(x) \sum_y P(y \ | \ x) = P(x)$$
which may be interpreted as a [discard condition](#operational-probabilistic-theories). (The
ordering is reflected by the fact that we can't derive $P(y)$ from
$\sum_{x} P(x,y) = \sum_x P(x) P(y \ | \ x)$.)
Let's consider some simple examples.
**Fork**
In the example of flu, sneezing and fever above, the graph has a *fork*
shape. For a probability distribution $P$ to satisfy the Markov
condition for this graph we must have
$$P(x, y, z) = P(x \ | \ z) P(y \ | \ z)P(z), \ \forall x,y,z.$$
However, $P(x,y) \neq P(x) P(y)$.
In other words, $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$, though $X$ and $Y$ are
not independent. This makes sense, we wouldn't expect sneezing and fever
to be uncorrelated, but given that we know whether or not Alice has flu,
telling us that she has fever isn't going to tell us anything about her
sneezing.
**Collider**
Reversing the arrows in the fork graph above gives a *collider* as in
the following example.
>
Moreover, intuitively we wouldn't expect there to be any other edges in
the above graph. Sneezing and fever, though correlated - each is more
likely if Alice has flu - are not direct causes of each other. That is,
$$P(sneezing \ | \ fever ) \neq P(sneezing) \ \text{ but } \ P(sneezing \ | \ fever, \ flu ) = P(sneezing \ | \ flu).$$
### *Bayesian networks*
Let $G$ be a *directed acyclic graph* or *DAG* $G$. (Here a directed
graph is a [presheaf on
($\bullet \rightrightarrows \bullet$)](https://golem.ph.utexas.edu/category/2008/01/mark_weber_on_nerves_of_catego.html)).
The set $Pa(Y)$ of *parents* of a node $Y$ of $G$ contains those nodes
$X$ of $G$ such that there is a directed edge $X \to Y$.
So, in the example above $Pa(flu) = \emptyset$ while
$Pa(fever) = Pa(sneezing) = \{ flu \}$.
To each node $X$ of a directed graph $G$, we may associate a random
variable, also denoted $X$. If $V$ is the set of nodes of $G$ and
$(x_X)_{X \in V}$ is a choice of value $x_X$ for each node $X$, such
that $y$ is the chosen value for $Y$, then $pa(y)$ will denote the
$Pa(Y)$-tuple of values $(x_X)_{X \in Pa(Y)}$.
To define Bayesian networks, and establish the notation, let's revise
some probability basics.
Let $P(x,y \ | \ z)$ mean $P(X = x \text{ and } \ Y = y \ | \ Z = z)$,
the *probability that $X$ has the value $x$, and $Y$ has the value $y$
given that $Z$ has the value $z$*. Recall that this is given by
$$P(x,y \ |\ z) = \frac{ P(x,y,z) }{P(z)}.$$
The *chain rule* says that, given a value $x$ of $X$ and sets of values
$\Omega, \Lambda$ of other random variables,
$$P(x, \Omega \ | \ \Lambda) = P( x \ | \ \Lambda) P( \Omega \ | \ x, \Lambda).$$
Random variables $X$ and $Y$ are said to be *conditionally independent
given $Z$*, written $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$, if for all values
$x$ of $X$, $y$ of $Y$ and $z$ of $Z$
$$P(x,y \ | \ z) = P(x \ | \ z) P(y \ | \ z).$$
By the chain rule this is equivalent to
$$P(x \ | \ y,z ) = P (x \ | \ z) , \ \forall x,y, z.$$
More generally, we may replace $X,Y$ and $Z$ with sets of random
variables. So, in the special case that $Z$ is empty, then $X$ and $Y$
are independent if and only if $P(x, y) = P(x)P(y)$ for all $x,y$.
#### Markov condition
A joint probability distribution $P$ on the nodes of a DAG $G$ is said
to satisfy the Markov condition if for any set of random
variable $\{X_i\}_{i = 1}^n$ on the nodes of $G$, with choice of values
$\{x_i\}_{i = 1}^n$
$$P(x_i, \dots, x_n) = \prod_{i = 1}^n P(x_i \ | \ {pa(x_i)}).$$
So, for the flu, fever and sneezing example above, a distribution $P$
satisfies the Markov condition if
$$P(flu, \ fever, \ sneezing) = P(fever \ | \ flu) P(sneezing \ | \ flu) P(flu).$$
A Bayesian network is defined as a pair $(G,P)$ of a DAG $G$ and a joint
probability distribution $P$ on the nodes of $G$ that satisfies the
Markov condition with respect to $G$. This means that each node in a
Bayesian network is conditionally independent, given its parents, of any
of the remaining nodes.
In particular, given a Bayesian network $(G,P)$ such that there is a
directed edge $X \to Y$, the Markov condition implies that
$$\sum_{y} P(x,y) = \sum_y P(x) P(y \ | \ x) = P(x) \sum_y P(y \ | \ x) = P(x)$$
which may be interpreted as a [discard condition](#operational-probabilistic-theories). (The
ordering is reflected by the fact that we can't derive $P(y)$ from
$\sum_{x} P(x,y) = \sum_x P(x) P(y \ | \ x)$.)
Let's consider some simple examples.
**Fork**
In the example of flu, sneezing and fever above, the graph has a *fork*
shape. For a probability distribution $P$ to satisfy the Markov
condition for this graph we must have
$$P(x, y, z) = P(x \ | \ z) P(y \ | \ z)P(z), \ \forall x,y,z.$$
However, $P(x,y) \neq P(x) P(y)$.
In other words, $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$, though $X$ and $Y$ are
not independent. This makes sense, we wouldn't expect sneezing and fever
to be uncorrelated, but given that we know whether or not Alice has flu,
telling us that she has fever isn't going to tell us anything about her
sneezing.
**Collider**
Reversing the arrows in the fork graph above gives a *collider* as in
the following example.
> 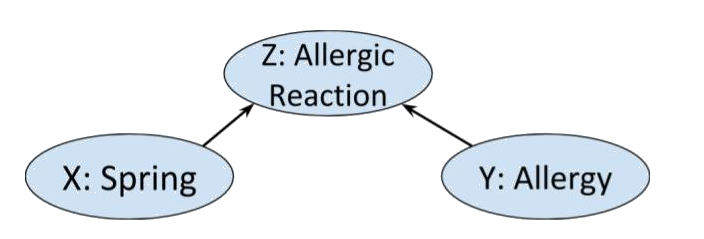 Clearly whether or not Alice has allergies other than hayfever is
independent of what season it is. So we'd expect a distribution on this
graph to satisfy $X \perp\!\!\!\!\!\!\!\perp Y \ | \ \emptyset$. However, if
we know that Alice is having an allergic reaction, and it happens to be spring, we will likely
assume that she has some allergy, i.e. $X$ and $Y$ are not conditionally independent given $Z$.
Indeed, the Markov condition and chain rule for this graph gives us $X \perp\!\!\!\!\!\!\!\perp Y \ | \ \emptyset$:
$$P(x, y, z) = P(x)P(y) P(z \ | \ x,\ y) = P(z \ | \ x,\ y) P( x\ | \ y) P(y) \ \forall x,y,z.$$
from which we cannot derive
$P(x \ | \ z) P(y \ | \ z) = P(x,y \ | \ z)$. (However, it could still
be true for some particular choice of probability distribution.)
**Chain**
Finally, let us return to the *chain* of correlations presented in the
introduction.
Clearly the probabilities that it is spring and that Alice is sneezing
are not independent, and indeed, we cannot derive $P(x, y) = P(x) P(y)$.
However observe that, by the chain rule, a Markov distribution on the
chain graph must satisfy $X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$. If we know
Alice is having an allergic reaction that is not hayfever, whether or
not she is sneezing is not going to affect our guess as to what season
it is.
Crucially, in this case, knowing the season is also not going to affect
whether we think Alice is sneezing. By definition, conditional
independence of $X$ and $Y$ given $Z$ is symmetric in $X$ and $Y$. In
other words, a joint distribution $P$ on the variables $X,Y,Z$ satisfies
the Markov condition with respect to the chain graph
$$X \longrightarrow Z \longrightarrow Y$$
if and only if $P$ satisfies
the Markov condition on
$$Y \longrightarrow Z \longrightarrow X .$$
### *d-separation*
The above observations can be generalised to statements about
conditional independences in any Bayesian network. That is, if $(G,P)$
is a Bayesian network then the structure of $G$ is enough to derive all
the conditional independences in $P$ that are implied by the graph $G$
(in reality there may be more that have not been included in the
network!).
Given a DAG $G$ and a set of vertices $U$ of $G$, let $m(U)$ denote the
union of $U$ with all the vertices $v$ of $G$ such that there is a
directed edge from $U$ to $v$. The set $W(U)$ will denote the
*non-inclusive future* of $U$, that is, the set of vertices $v$ of $G$
for which there is no directed (possibly trivial) path from $v$ to $U$.
For a graph $G$, let $X, Y, Z$ now denote disjoint subsets of the
vertices of $G$ (and their corresponding random variables). Set
$W := W(X \cup Y \cup Z)$.
Then $X$ and $Y$ are said to be *d-separated* by $Z$, written
$X \perp Y \ | \ Z$, if there is a partition $\{U,V,W,Z\}$ of the nodes
of $G$ such that
- $X \subseteq U$ and $Y \subseteq V$, and
- $m(U) \cap m(V) \subseteq W,$ in other words $U$ and $V$ have no
direct influence on each other.
(This is lemma 19 in the paper.)
Now d-separation is really useful since it tells us everything there is
to know about the conditional dependences on Bayesian networks with
underlying graph $G$. Indeed,
#### Theorem 5
- **Soundness of d-separation** ([Verma and Pearl,
1988](https://www.sciencedirect.com/science/article/pii/B9780444886507500111))
If $P$ is a Markov distribution with respect to a graph $G$ then for
all disjoint subsets $X,Y,Z$ of nodes of $G$ $X \perp Y \ | \ Z$
implies that $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$.
- **Completeness of d-separation** ([Meek,
1995](https://dl.acm.org/citation.cfm?id=2074205)) If
$X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$ for all $P$ Markov with respect to
$G$, then $X \perp Y \ | \ Z$.
We can combine the previous examples of fork, collider and chain graphs
to get the following
>
Clearly whether or not Alice has allergies other than hayfever is
independent of what season it is. So we'd expect a distribution on this
graph to satisfy $X \perp\!\!\!\!\!\!\!\perp Y \ | \ \emptyset$. However, if
we know that Alice is having an allergic reaction, and it happens to be spring, we will likely
assume that she has some allergy, i.e. $X$ and $Y$ are not conditionally independent given $Z$.
Indeed, the Markov condition and chain rule for this graph gives us $X \perp\!\!\!\!\!\!\!\perp Y \ | \ \emptyset$:
$$P(x, y, z) = P(x)P(y) P(z \ | \ x,\ y) = P(z \ | \ x,\ y) P( x\ | \ y) P(y) \ \forall x,y,z.$$
from which we cannot derive
$P(x \ | \ z) P(y \ | \ z) = P(x,y \ | \ z)$. (However, it could still
be true for some particular choice of probability distribution.)
**Chain**
Finally, let us return to the *chain* of correlations presented in the
introduction.
Clearly the probabilities that it is spring and that Alice is sneezing
are not independent, and indeed, we cannot derive $P(x, y) = P(x) P(y)$.
However observe that, by the chain rule, a Markov distribution on the
chain graph must satisfy $X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$. If we know
Alice is having an allergic reaction that is not hayfever, whether or
not she is sneezing is not going to affect our guess as to what season
it is.
Crucially, in this case, knowing the season is also not going to affect
whether we think Alice is sneezing. By definition, conditional
independence of $X$ and $Y$ given $Z$ is symmetric in $X$ and $Y$. In
other words, a joint distribution $P$ on the variables $X,Y,Z$ satisfies
the Markov condition with respect to the chain graph
$$X \longrightarrow Z \longrightarrow Y$$
if and only if $P$ satisfies
the Markov condition on
$$Y \longrightarrow Z \longrightarrow X .$$
### *d-separation*
The above observations can be generalised to statements about
conditional independences in any Bayesian network. That is, if $(G,P)$
is a Bayesian network then the structure of $G$ is enough to derive all
the conditional independences in $P$ that are implied by the graph $G$
(in reality there may be more that have not been included in the
network!).
Given a DAG $G$ and a set of vertices $U$ of $G$, let $m(U)$ denote the
union of $U$ with all the vertices $v$ of $G$ such that there is a
directed edge from $U$ to $v$. The set $W(U)$ will denote the
*non-inclusive future* of $U$, that is, the set of vertices $v$ of $G$
for which there is no directed (possibly trivial) path from $v$ to $U$.
For a graph $G$, let $X, Y, Z$ now denote disjoint subsets of the
vertices of $G$ (and their corresponding random variables). Set
$W := W(X \cup Y \cup Z)$.
Then $X$ and $Y$ are said to be *d-separated* by $Z$, written
$X \perp Y \ | \ Z$, if there is a partition $\{U,V,W,Z\}$ of the nodes
of $G$ such that
- $X \subseteq U$ and $Y \subseteq V$, and
- $m(U) \cap m(V) \subseteq W,$ in other words $U$ and $V$ have no
direct influence on each other.
(This is lemma 19 in the paper.)
Now d-separation is really useful since it tells us everything there is
to know about the conditional dependences on Bayesian networks with
underlying graph $G$. Indeed,
#### Theorem 5
- **Soundness of d-separation** ([Verma and Pearl,
1988](https://www.sciencedirect.com/science/article/pii/B9780444886507500111))
If $P$ is a Markov distribution with respect to a graph $G$ then for
all disjoint subsets $X,Y,Z$ of nodes of $G$ $X \perp Y \ | \ Z$
implies that $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$.
- **Completeness of d-separation** ([Meek,
1995](https://dl.acm.org/citation.cfm?id=2074205)) If
$X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$ for all $P$ Markov with respect to
$G$, then $X \perp Y \ | \ Z$.
We can combine the previous examples of fork, collider and chain graphs
to get the following
> 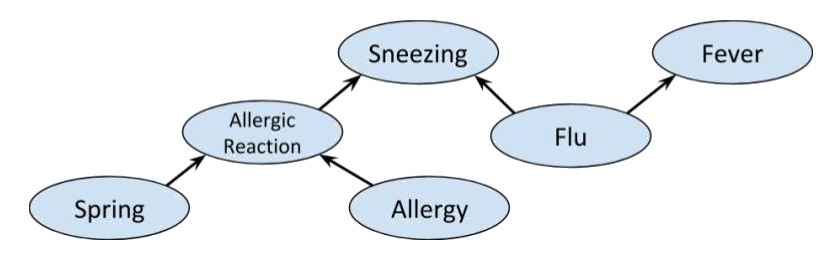 A priori, *Allergic reaction* is conditionally independent of *Fever*.
Indeed, we have the partition
>
A priori, *Allergic reaction* is conditionally independent of *Fever*.
Indeed, we have the partition
> 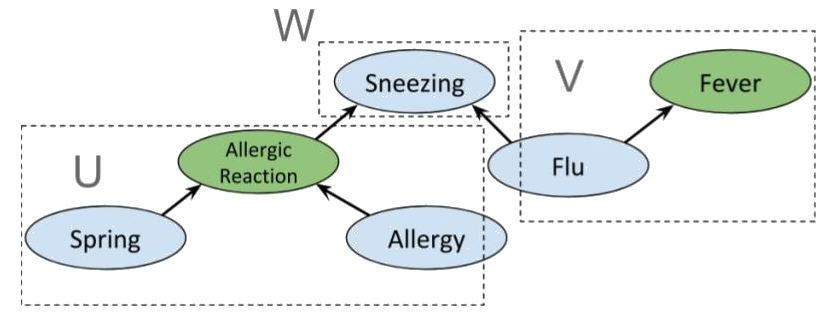 which clearly satisfies d-separation. However, if *Sneezing* is known then $W = \emptyset$,
so *Allergic reaction* and *Fever* are not independent. Indeed, if we use the same sets $U$ and $V$ as before, then
$m(U) \cap m(V) = \{ Sneezing \}$, so the condition for d-separation fails; and it does for any possible choice of $U$ and $V$.
Interestingly, if *Flu* is also known, we again obtain conditional independence between *Allergic reaction* and *Fever*, as shown below.
>
which clearly satisfies d-separation. However, if *Sneezing* is known then $W = \emptyset$,
so *Allergic reaction* and *Fever* are not independent. Indeed, if we use the same sets $U$ and $V$ as before, then
$m(U) \cap m(V) = \{ Sneezing \}$, so the condition for d-separation fails; and it does for any possible choice of $U$ and $V$.
Interestingly, if *Flu* is also known, we again obtain conditional independence between *Allergic reaction* and *Fever*, as shown below.
> 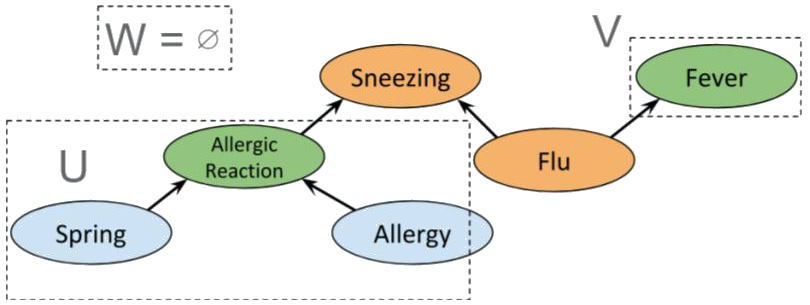 Before describing the limitations of this setup and why we may want to
generalise it, it is worth observing that Theorem 5 is genuinely useful
computationally. Theorem 5 says that given a Bayesian network $(G,P)$,
the structure of $G$ gives us a recipe to factor $P$, thereby greatly
increasing computation efficiency for Bayesian inference.
### *Latent variables, hidden variables, and unobservables*
In the context of Bayesian networks, there are two reasons that we may
wish to add variables to a probabilistic model, even if we are not
entirely sure what the variables signify or how they are distributed.
The first reason is statistical and the second is physical.
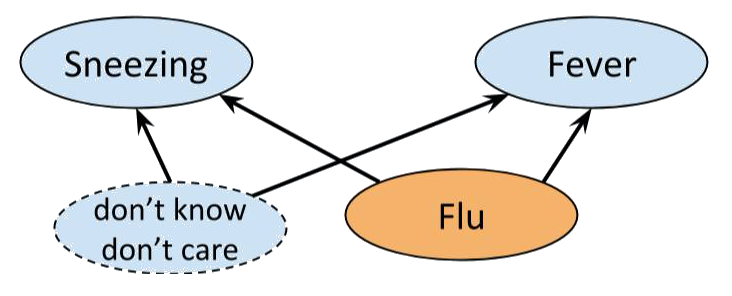
Consider the example of flu, fever and sneezing discussed earlier. Although our analysis told us $Fever \perp\!\!\!\!\!\!\!\perp Sneezing \ | \ Flu$, if we conduct an experiment we are likely to find:
$$P(fever \ | \ sneezing, \ flu) \neq P(fever \ | \ flu).$$
The problem is caused by the graph not properly modelling reality, but a simplification of it.
After all, there are a whole bunch of things that can cause sneezing and
flu. We just don't know what they all are or how to measure them. So, to
make the network work, we may add a hypothetical *latent variable* that
bunches together all the unknown joint causes, and equip it with a
distribution that makes the whole network Bayesian, so that we are still
able to perform inference methods like belief propagation.
>
Before describing the limitations of this setup and why we may want to
generalise it, it is worth observing that Theorem 5 is genuinely useful
computationally. Theorem 5 says that given a Bayesian network $(G,P)$,
the structure of $G$ gives us a recipe to factor $P$, thereby greatly
increasing computation efficiency for Bayesian inference.
### *Latent variables, hidden variables, and unobservables*
In the context of Bayesian networks, there are two reasons that we may
wish to add variables to a probabilistic model, even if we are not
entirely sure what the variables signify or how they are distributed.
The first reason is statistical and the second is physical.
Consider the example of flu, fever and sneezing discussed earlier. Although our analysis told us $Fever \perp\!\!\!\!\!\!\!\perp Sneezing \ | \ Flu$, if we conduct an experiment we are likely to find:
$$P(fever \ | \ sneezing, \ flu) \neq P(fever \ | \ flu).$$
The problem is caused by the graph not properly modelling reality, but a simplification of it.
After all, there are a whole bunch of things that can cause sneezing and
flu. We just don't know what they all are or how to measure them. So, to
make the network work, we may add a hypothetical *latent variable* that
bunches together all the unknown joint causes, and equip it with a
distribution that makes the whole network Bayesian, so that we are still
able to perform inference methods like belief propagation.
>  On the other hand, we may want to add variables to a Bayesian network if
we have evidence that doing so will provide a better model of reality.
For example, consider the network with just two connected nodes
>
On the other hand, we may want to add variables to a Bayesian network if
we have evidence that doing so will provide a better model of reality.
For example, consider the network with just two connected nodes
> 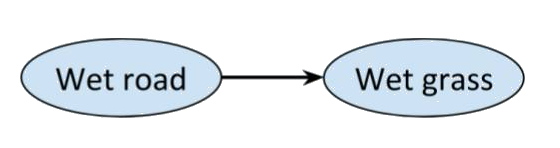 Every distribution on this graph is Markov, and we would expect there to
be a correlation between a road being wet and the grass next to it being
wet as well, but most people would claim that there's something missing
from the picture. After all, rain could be a 'common cause' of the road
and the grass being wet. So, it makes sense to add a third variable.
But maybe we can't observe whether it has rained or not, only whether
the grass and/or road are wet. Nonetheless, the correlation we observe
suggests that they have a common cause. To deal with such cases, we
could make the third variable *hidden*. We may not know what information
is included in a hidden variable, nor its probability distribution.
All that matters is that the hidden variable helps to explain the
observed correlations.
>
Every distribution on this graph is Markov, and we would expect there to
be a correlation between a road being wet and the grass next to it being
wet as well, but most people would claim that there's something missing
from the picture. After all, rain could be a 'common cause' of the road
and the grass being wet. So, it makes sense to add a third variable.
But maybe we can't observe whether it has rained or not, only whether
the grass and/or road are wet. Nonetheless, the correlation we observe
suggests that they have a common cause. To deal with such cases, we
could make the third variable *hidden*. We may not know what information
is included in a hidden variable, nor its probability distribution.
All that matters is that the hidden variable helps to explain the
observed correlations.
>  So, latent variables are a statistical tool that ensure the Markov
condition holds. Hence they are inherently classical, and can, in
theory, be known. But the universe is not classical, so, even if we lump
whatever we want into as many classical hidden variables as we want and
put them wherever we need, in some cases, there will still be
empirically observed correlations that do not satisfy the Markov
condition.
Most famously, Bell's experiment shows that it is possible to have
distinct variables $A$ and $B$ that exhibit correlations that cannot be
explained by any classical hidden variable, since classical variables
are restricted by the principle of locality.
In other words, though $A \perp B \ | \ \Lambda$,
$$P(a \ | b,\ \lambda) \neq P(a \ | \ \lambda).$$
Implicitly, this means that a *classical* $\Lambda$ is not enough. If we
want $P(a \ | b,\ \lambda) \neq P(a \ | \ \lambda)$ to hold, $\Lambda$
must be a non-local (non-classical) variable. Quantum mechanics implies
that we can't possibly empirically find the value of a non-local
variable (for similar reasons to the Heisenberg's uncertainty
principle), so non-classical variables are often called *unobservables*.
In particular, it is irrelevant to question whether
$A \perp\!\!\!\!\!\!\!\perp B \ | \ \Lambda$, as we would need to know the value
of $\Lambda$ in order to condition over it.
Indeed, this is the key idea behind what follows. We declare certain
variables to be unobservable and then insist that conditional (in)dependence
only makes sense between *observable variables conditioned
over observable variables*.
## **Generalising classical causality**
The correlations observed in the Bell experiment can be explained by
quantum mechanics. But thought experiments such as the one described
[here](https://www.nature.com/articles/nphys2916) suggest that
theoretically, correlations may exist that violate even quantum
causality.
So, given that graphical models and d-separation provide such a powerful
tool for causal reasoning in the classical context, how can we
generalise the Markov condition and Theorem 5 to quantum, and even more
general causal theories? And, if we have a *theory-independent* Markov
condition, are there d-separation results that don't correspond to any
given causal theory?
Clearly the first step in answering these questions is to fix a
definition of a *causal* theory.
### *Operational probabalistic theories*
An [*operational theory*](https://arxiv.org/pdf/0908.1583.pdf) is a
symmetric monoidal category $(\mathsf {C}, \otimes, I)$ whose objects are
known as *systems* or *resources*. Morphisms are finite sets
$f = \{\mathcal {C}_i\}_{i \in I}$ called *tests*, whose elements are
called *outcomes*. Tests with a single element are called
*deterministic*, and for each system $A \in ob (\mathsf {C})$, the
identity $id_A \in \mathsf (A,A)$ is a deterministic test.
In this discussion, we'll identify tests
$\{\mathcal {C}_i \}_i , \{\mathcal {D}_j\}_j$ in $\mathsf {C}$ if we may
always replace one with the other without affecting the distributions in
$\mathsf {C}(I, I)$.
Given $\{\mathcal {C}_i \}_i \in \mathsf {C}(B, C)$ and
$\{\mathcal {D}_j \} \in \mathsf {C}(A, B)$, their composition $f \circ g$
is given by
$$\{ \mathcal {C}_i \circ \mathcal {D}_j \}_{i,j} \in \mathsf {C}(A, C).$$
First apply $\mathcal {D}$ with output $B$ then apply $\mathcal {C}$ with
outcome $C$.
The monoidal composition
$\{ \mathcal {C}_i \otimes \mathcal {D}_j \}_{i, j} \in \mathsf {C}(A \otimes C, B \otimes D)$
corresponds to applying $\{\mathcal {C}_i\}_i \in \mathsf {C}(A,B)$ and
$\{ \mathcal {D}_j \}_j$ separately on $A$ and $C$.
An *operational probabilistic theory* or *OPT* is an operational theory
such that every test $I \to I$ is a probability distribution.
A morphism $\{ \mathcal {C}_i \}_i \in \mathsf {C}(A, I)$ is called an
*effect* on $A$. An OPT $\mathsf {C}$ is called *causal* or a *causal
theory* if, for each system $A \in ob (\mathsf {C})$, there is a unique
deterministic effect $\top_A \in \mathsf {C}( A, I)$ which we call the
*discard* of $A$.
In particular, for a causal OPT $\mathsf {C}$, uniqueness of the discard
implies that, for all systems $A, B \in ob (\mathsf {C})$,
$$\top_A \otimes \top_B = \top_{A \otimes B},$$
and, given any
determinstic test $\mathcal {C} \in \mathsf {C}(A, B)$,
$$\top_B \circ \mathcal {C} = \top_A.$$
The existence of a discard map allows a definition of *causal morphisms*
in a causal theory. For example, as we saw in
[January](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
when we discussed [Kissinger and Uijlen's
paper](https://arxiv.org/abs/1701.04732), a test
$\{ \mathcal {C}_i \}_i \in \mathsf {C} (A, B)$ is *causal* if
$$\top_B \circ \{ \mathcal {C}_i \}_i = \top_A \in \mathsf {C}( A, I).$$
In other words, for a causal test, discarding the outcome is the same as
not performing the test. Intuitively it is not obvious why such
morphisms should be called causal. But this definition enables the
formulation of a [*non-signalling
condition*](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
that describes the conditions under which the possibility of
cause-effect correlation is excluded, in particular, it implies the
impossibility of time travel.
#### Examples
The category
[$Mat(\mathbb {R}_+)$](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
of natural numbers and with $Mat(\mathbb {R}_+)(m,n)$ the set of
$n \times m$ matrices, has the structure of a causal OPT. The causal
morphisms in $Mat(\mathbb {R}_+)$ are the stochastic maps (the matrices
whose columns sum to 1). This category describes classical probability
theory.
The category [$\mathsf{CPM}$](https://arxiv.org/abs/1701.04732) of sets
of linear operators on Hilbert spaces and completely positive maps
between them is an OPT and describes quantum relations. The causal
morphisms are the trace preserving completely positive maps.
Finally, [Boxworld](https://www.nature.com/articles/nphys2916) is the
theory that allows to describe any correlation between two
variables as the cause of some resource of the theory in the past.
### *Generalised Bayesian networks*
So, we're finally ready to give the main construction and results of the
paper. As mentioned before, to get a generalised d-separation result,
the idea is that we will distinguish observable and unobservable
variables, and simply insist that conditional independence is only
defined relative to observable variables.
To this end, a *generalised DAG* or *GDAG* is a DAG $G$ together with a
partition on the nodes of $G$ into two subsets called *observed* and
*unobserved*. We'll represent observed nodes by triangles, and
unobserved nodes by circles. An edge out of an (un)observed node will be
called *(un)observed* and represented by a (solid) dashed arrow.
In order to get a generalisation of Theorem 5, we still need to come up
with a sensible generalisation of the Markov property which will
essentially say that at an observed node that has only observed parents,
the distribution must be Markov. However, if an observed node has an
unobserved parent, the latter's whole history is needed to describe the
distribution.
To state this precisely, we will associate a causal theory
$(\mathsf {C}, \otimes, I)$ to a GDAG $G$ via an assignment of systems to
edges of $G$ and tests to nodes of $G$, such that the observed edges of
$G$ will 'carry' only the outcomes of classical tests (so will say
something about conditional probability) whereas unobserved edges will
carry only the output system.
Precisely, such an assignment $P$ satisfies the *generalised Markov
condition (GMC)* and is called a *generalised Markov distribution* if
- Each unobserved edge corresponds to a distinct system in the theory.
- *If we can't observe what is happening at a node, we can't condition
over it:* To each unobserved node and each value of its observed
parents, we assign a deterministic test from the system defined by
the product of its incoming (unobserved) edges to the system defined
by the product of its outgoing (unobserved) edges.
- Each observed node $X$ is an observation test, i.e. a morphism in
$\mathsf {C}(A, I)$ for the system $A \in ob( \mathsf {C})$
corresponding to the product of the systems assigned to the
unobserved input edges of $X$. Since $\mathsf {C}$ is a causal theory,
this says that $X$ is assigned a classical random variable, also
denoted $X$, and that if $Y$ is an observed node, and has observed
parent $X$, the distribution at $Y$ is conditionally dependent on
the distribution at $X$ (see
[here](https://arxiv.org/pdf/0908.1583.pdf) for details).
- It therefore follows that each observed edge is assigned the trivial system $I$.
- The joint probability distribution on the observed nodes of $G$ is given by the morphism $\mathsf {C}(I, I)$
that results from these assignments.
A *generalised Bayesian network* consists of a GDAG $G$ together with a
generalised Markov distribution $P$ on $G$.
#### Example
Consider the following GDAG
>
So, latent variables are a statistical tool that ensure the Markov
condition holds. Hence they are inherently classical, and can, in
theory, be known. But the universe is not classical, so, even if we lump
whatever we want into as many classical hidden variables as we want and
put them wherever we need, in some cases, there will still be
empirically observed correlations that do not satisfy the Markov
condition.
Most famously, Bell's experiment shows that it is possible to have
distinct variables $A$ and $B$ that exhibit correlations that cannot be
explained by any classical hidden variable, since classical variables
are restricted by the principle of locality.
In other words, though $A \perp B \ | \ \Lambda$,
$$P(a \ | b,\ \lambda) \neq P(a \ | \ \lambda).$$
Implicitly, this means that a *classical* $\Lambda$ is not enough. If we
want $P(a \ | b,\ \lambda) \neq P(a \ | \ \lambda)$ to hold, $\Lambda$
must be a non-local (non-classical) variable. Quantum mechanics implies
that we can't possibly empirically find the value of a non-local
variable (for similar reasons to the Heisenberg's uncertainty
principle), so non-classical variables are often called *unobservables*.
In particular, it is irrelevant to question whether
$A \perp\!\!\!\!\!\!\!\perp B \ | \ \Lambda$, as we would need to know the value
of $\Lambda$ in order to condition over it.
Indeed, this is the key idea behind what follows. We declare certain
variables to be unobservable and then insist that conditional (in)dependence
only makes sense between *observable variables conditioned
over observable variables*.
## **Generalising classical causality**
The correlations observed in the Bell experiment can be explained by
quantum mechanics. But thought experiments such as the one described
[here](https://www.nature.com/articles/nphys2916) suggest that
theoretically, correlations may exist that violate even quantum
causality.
So, given that graphical models and d-separation provide such a powerful
tool for causal reasoning in the classical context, how can we
generalise the Markov condition and Theorem 5 to quantum, and even more
general causal theories? And, if we have a *theory-independent* Markov
condition, are there d-separation results that don't correspond to any
given causal theory?
Clearly the first step in answering these questions is to fix a
definition of a *causal* theory.
### *Operational probabalistic theories*
An [*operational theory*](https://arxiv.org/pdf/0908.1583.pdf) is a
symmetric monoidal category $(\mathsf {C}, \otimes, I)$ whose objects are
known as *systems* or *resources*. Morphisms are finite sets
$f = \{\mathcal {C}_i\}_{i \in I}$ called *tests*, whose elements are
called *outcomes*. Tests with a single element are called
*deterministic*, and for each system $A \in ob (\mathsf {C})$, the
identity $id_A \in \mathsf (A,A)$ is a deterministic test.
In this discussion, we'll identify tests
$\{\mathcal {C}_i \}_i , \{\mathcal {D}_j\}_j$ in $\mathsf {C}$ if we may
always replace one with the other without affecting the distributions in
$\mathsf {C}(I, I)$.
Given $\{\mathcal {C}_i \}_i \in \mathsf {C}(B, C)$ and
$\{\mathcal {D}_j \} \in \mathsf {C}(A, B)$, their composition $f \circ g$
is given by
$$\{ \mathcal {C}_i \circ \mathcal {D}_j \}_{i,j} \in \mathsf {C}(A, C).$$
First apply $\mathcal {D}$ with output $B$ then apply $\mathcal {C}$ with
outcome $C$.
The monoidal composition
$\{ \mathcal {C}_i \otimes \mathcal {D}_j \}_{i, j} \in \mathsf {C}(A \otimes C, B \otimes D)$
corresponds to applying $\{\mathcal {C}_i\}_i \in \mathsf {C}(A,B)$ and
$\{ \mathcal {D}_j \}_j$ separately on $A$ and $C$.
An *operational probabilistic theory* or *OPT* is an operational theory
such that every test $I \to I$ is a probability distribution.
A morphism $\{ \mathcal {C}_i \}_i \in \mathsf {C}(A, I)$ is called an
*effect* on $A$. An OPT $\mathsf {C}$ is called *causal* or a *causal
theory* if, for each system $A \in ob (\mathsf {C})$, there is a unique
deterministic effect $\top_A \in \mathsf {C}( A, I)$ which we call the
*discard* of $A$.
In particular, for a causal OPT $\mathsf {C}$, uniqueness of the discard
implies that, for all systems $A, B \in ob (\mathsf {C})$,
$$\top_A \otimes \top_B = \top_{A \otimes B},$$
and, given any
determinstic test $\mathcal {C} \in \mathsf {C}(A, B)$,
$$\top_B \circ \mathcal {C} = \top_A.$$
The existence of a discard map allows a definition of *causal morphisms*
in a causal theory. For example, as we saw in
[January](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
when we discussed [Kissinger and Uijlen's
paper](https://arxiv.org/abs/1701.04732), a test
$\{ \mathcal {C}_i \}_i \in \mathsf {C} (A, B)$ is *causal* if
$$\top_B \circ \{ \mathcal {C}_i \}_i = \top_A \in \mathsf {C}( A, I).$$
In other words, for a causal test, discarding the outcome is the same as
not performing the test. Intuitively it is not obvious why such
morphisms should be called causal. But this definition enables the
formulation of a [*non-signalling
condition*](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
that describes the conditions under which the possibility of
cause-effect correlation is excluded, in particular, it implies the
impossibility of time travel.
#### Examples
The category
[$Mat(\mathbb {R}_+)$](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
of natural numbers and with $Mat(\mathbb {R}_+)(m,n)$ the set of
$n \times m$ matrices, has the structure of a causal OPT. The causal
morphisms in $Mat(\mathbb {R}_+)$ are the stochastic maps (the matrices
whose columns sum to 1). This category describes classical probability
theory.
The category [$\mathsf{CPM}$](https://arxiv.org/abs/1701.04732) of sets
of linear operators on Hilbert spaces and completely positive maps
between them is an OPT and describes quantum relations. The causal
morphisms are the trace preserving completely positive maps.
Finally, [Boxworld](https://www.nature.com/articles/nphys2916) is the
theory that allows to describe any correlation between two
variables as the cause of some resource of the theory in the past.
### *Generalised Bayesian networks*
So, we're finally ready to give the main construction and results of the
paper. As mentioned before, to get a generalised d-separation result,
the idea is that we will distinguish observable and unobservable
variables, and simply insist that conditional independence is only
defined relative to observable variables.
To this end, a *generalised DAG* or *GDAG* is a DAG $G$ together with a
partition on the nodes of $G$ into two subsets called *observed* and
*unobserved*. We'll represent observed nodes by triangles, and
unobserved nodes by circles. An edge out of an (un)observed node will be
called *(un)observed* and represented by a (solid) dashed arrow.
In order to get a generalisation of Theorem 5, we still need to come up
with a sensible generalisation of the Markov property which will
essentially say that at an observed node that has only observed parents,
the distribution must be Markov. However, if an observed node has an
unobserved parent, the latter's whole history is needed to describe the
distribution.
To state this precisely, we will associate a causal theory
$(\mathsf {C}, \otimes, I)$ to a GDAG $G$ via an assignment of systems to
edges of $G$ and tests to nodes of $G$, such that the observed edges of
$G$ will 'carry' only the outcomes of classical tests (so will say
something about conditional probability) whereas unobserved edges will
carry only the output system.
Precisely, such an assignment $P$ satisfies the *generalised Markov
condition (GMC)* and is called a *generalised Markov distribution* if
- Each unobserved edge corresponds to a distinct system in the theory.
- *If we can't observe what is happening at a node, we can't condition
over it:* To each unobserved node and each value of its observed
parents, we assign a deterministic test from the system defined by
the product of its incoming (unobserved) edges to the system defined
by the product of its outgoing (unobserved) edges.
- Each observed node $X$ is an observation test, i.e. a morphism in
$\mathsf {C}(A, I)$ for the system $A \in ob( \mathsf {C})$
corresponding to the product of the systems assigned to the
unobserved input edges of $X$. Since $\mathsf {C}$ is a causal theory,
this says that $X$ is assigned a classical random variable, also
denoted $X$, and that if $Y$ is an observed node, and has observed
parent $X$, the distribution at $Y$ is conditionally dependent on
the distribution at $X$ (see
[here](https://arxiv.org/pdf/0908.1583.pdf) for details).
- It therefore follows that each observed edge is assigned the trivial system $I$.
- The joint probability distribution on the observed nodes of $G$ is given by the morphism $\mathsf {C}(I, I)$
that results from these assignments.
A *generalised Bayesian network* consists of a GDAG $G$ together with a
generalised Markov distribution $P$ on $G$.
#### Example
Consider the following GDAG
> 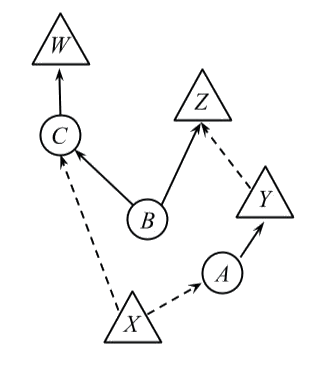 Let's build its OPT morphism as indictated by the *generalised Markov condition*.
The observed node $X$ has no incoming edges so it corresponds to a $\mathsf {C}(I, I)$ morphism, and thus we assign a probability
distribution to it.
The unobserved node A *depends* on $X$, and has no unobserved inputs, so
we assign a deterministic test $A(x): I \to A$ for each value $x$ of
$X$.
>
Let's build its OPT morphism as indictated by the *generalised Markov condition*.
The observed node $X$ has no incoming edges so it corresponds to a $\mathsf {C}(I, I)$ morphism, and thus we assign a probability
distribution to it.
The unobserved node A *depends* on $X$, and has no unobserved inputs, so
we assign a deterministic test $A(x): I \to A$ for each value $x$ of
$X$.
> 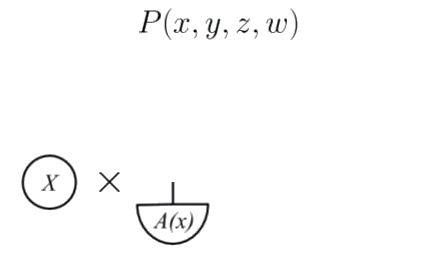 The observed node $Y$ has one incoming unobserved edge and no incoming
observed edges so we assign to it a test $Y: A \to I$ such that, for
each value $x$ of $X$, $Y \circ A(x)$ is a probability distribution.
Building up the rest of the picture gives an OPT diagram of the form
>
The observed node $Y$ has one incoming unobserved edge and no incoming
observed edges so we assign to it a test $Y: A \to I$ such that, for
each value $x$ of $X$, $Y \circ A(x)$ is a probability distribution.
Building up the rest of the picture gives an OPT diagram of the form
> 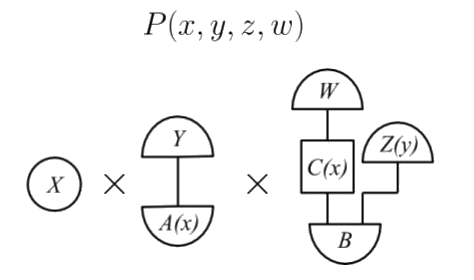 which is a $\mathsf {C}(I, I)$ morphism that defines the joint probability distribution $P(x,y,z,w)$.
We now have all the ingredients to state Theorem 22, the *generalised
d-separation theorem*. This is the analogue of Theorem 5 for generalised
Markov distributions.
#### Theorem 22
Given a GDAG $G$ and subsets $X,Y, Z$ of observed nodes
- if a probability distribution $P$ is generalised Markov relative to
$G$ then
$X \perp Y \ | \ Z \Rightarrow X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$.
- If $X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$ holds for all generalised Markov
probability distributions on $G$, then $X \perp Y \ | \ Z$.
Note in particular that there is no change in the definition of
d-separation: d-separation of a GDAG $G$ is simply d-separation with
respect to its underlying DAG. There is also no change in the definition
of conditional independence. Now, however, we restrict to statements of
conditional independence with respect to observed nodes only. This
enables the generalised soundness and completeness statements of the
theorem.
The proof of soundness uses uniqueness of discarding, and completeness
follows since generalised Markov is a stronger condition on a
distribution than classically Markov.
### *Classical distributions on GDAGs*
Theorem 22 is all well and good. But does it really generalise the
classical case? That is, can we recover Theorem 5 for all classical
Bayesian networks from Theorem 22?
As a first step, Proposition 17 states that if all the nodes of a
generalised Bayesian network are observed, then it is a classical
bayesian network. In fact, this follows pretty immediately from the
definitions.
Moreover, it is easily checked that, given a classical Bayesian network,
even if it has hidden or latent variables, it can still be expressed
directly as a generalised Bayesian network with no unobserved nodes.
In fact, Theorem 22 generalises Theorem 5 in a stricter sense. That is,
the generalised Bayesian network setup together with classical causality
adds nothing extra to the theory of classical Bayesian networks. If a
generalised Markov distribution is classical (then hidden and latent
variables may be represented by unobserved nodes), it can be viewed as a
classical Bayesian network. More precisely, Lemma 18 says that, given
any generalised Bayesian network $(G,P)$ with underlying DAG $G'$ and
distribution $P \in \mathcal {C}$, we can construct a classical Bayesian
network $(G', P')$ such that $P'$ agrees with $P$ on the observed nodes.
It is worth voicing a note of caution. The authors themselves
mention in the conclusion that the construction based on GDAGs with two
types of nodes is not entirely satisfactory. The problem is that,
although the setups and results presented here do give a generalisation
of Theorem 5, they do not, as such, provide a way of generalising
Bayesian networks as they are used for probabalistic inference to
non-classical settings. For example, belief propagation works through
observed nodes, but there is no apparent way of generalising it for
unobserved nodes.
## **Theory independence**
More generally, given a GDAG $G$, we can look at the set of
distributions on $G$ that are generalised Markov with respect to a given
causal theory. Of particular importance are the following.
- The set $\mathcal {C}$ of generalised Markov distributions in
$Mat(\mathbb {R}_+)$ on $G$.
- The set $\mathcal {Q}$ of generalised Markov distributions in
$\mathsf{CPM}$ on $G$.
- The set $\mathcal {G}$ of all generalised Markov distributions on $G$.
(This is the set of generalised Markov distributions in
[Boxworld](https://www.nature.com/articles/nphys2916).)
Moreover, we can distinguish another class of distributions on $G$, by
not restricting to d-seperation of observed nodes, but considering
distributions that satisfy the observable conditional independences
given by any d-separation properties on the graph. Theorem 22 implies,
in particular that $G \subseteq I$.
And, so, since $Mat(\mathbb {R}_+)$ embeds into $\mathsf{CPM}$, we have
$\mathcal {C} \subseteq \mathcal {Q} \subseteq \mathcal {G} \subseteq \mathcal {I}$.
This means that one can ask for which graphs (some or all of) these
inequalities are strict, and the last part of the paper explores these
questions. In the [original paper](http://iopscience.iop.org/article/10.1088/1367-2630/16/11/113043/meta),
a sufficient condition is given for graphs to
satisfy $\mathcal {C} \neq \mathcal {I}$. I.e. for these graphs it is
guaranteed that the causal structure admits correlations that are
non-local. Moreover the authors show that their condition is necessary for
small enough graphs.
Another interesting result is that there exist graphs for which
$\mathcal {G} \neq \mathcal {I}$. This means that using a theory of
resources, whatever theory it may be, to explain correlations imposes
constraints that are stronger than those imposed by the relations
themselves.
## **What next?**
This setup represents one direction for using category theory to
generalise Bayesian networks. In our group work at the ACT workshop, we
considered another generalisation of Bayesian networks, this time
staying within the classical realm. Namely, building on the work of
[Bonchi, Gadducci, Kissinger, Sobocinski, and
Zanasi](https://arxiv.org/abs/1602.06771), we gave a functorial Markov
condition on directed graphs admitting cycles. Hopefully we'll present
this work here soon.
which is a $\mathsf {C}(I, I)$ morphism that defines the joint probability distribution $P(x,y,z,w)$.
We now have all the ingredients to state Theorem 22, the *generalised
d-separation theorem*. This is the analogue of Theorem 5 for generalised
Markov distributions.
#### Theorem 22
Given a GDAG $G$ and subsets $X,Y, Z$ of observed nodes
- if a probability distribution $P$ is generalised Markov relative to
$G$ then
$X \perp Y \ | \ Z \Rightarrow X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$.
- If $X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$ holds for all generalised Markov
probability distributions on $G$, then $X \perp Y \ | \ Z$.
Note in particular that there is no change in the definition of
d-separation: d-separation of a GDAG $G$ is simply d-separation with
respect to its underlying DAG. There is also no change in the definition
of conditional independence. Now, however, we restrict to statements of
conditional independence with respect to observed nodes only. This
enables the generalised soundness and completeness statements of the
theorem.
The proof of soundness uses uniqueness of discarding, and completeness
follows since generalised Markov is a stronger condition on a
distribution than classically Markov.
### *Classical distributions on GDAGs*
Theorem 22 is all well and good. But does it really generalise the
classical case? That is, can we recover Theorem 5 for all classical
Bayesian networks from Theorem 22?
As a first step, Proposition 17 states that if all the nodes of a
generalised Bayesian network are observed, then it is a classical
bayesian network. In fact, this follows pretty immediately from the
definitions.
Moreover, it is easily checked that, given a classical Bayesian network,
even if it has hidden or latent variables, it can still be expressed
directly as a generalised Bayesian network with no unobserved nodes.
In fact, Theorem 22 generalises Theorem 5 in a stricter sense. That is,
the generalised Bayesian network setup together with classical causality
adds nothing extra to the theory of classical Bayesian networks. If a
generalised Markov distribution is classical (then hidden and latent
variables may be represented by unobserved nodes), it can be viewed as a
classical Bayesian network. More precisely, Lemma 18 says that, given
any generalised Bayesian network $(G,P)$ with underlying DAG $G'$ and
distribution $P \in \mathcal {C}$, we can construct a classical Bayesian
network $(G', P')$ such that $P'$ agrees with $P$ on the observed nodes.
It is worth voicing a note of caution. The authors themselves
mention in the conclusion that the construction based on GDAGs with two
types of nodes is not entirely satisfactory. The problem is that,
although the setups and results presented here do give a generalisation
of Theorem 5, they do not, as such, provide a way of generalising
Bayesian networks as they are used for probabalistic inference to
non-classical settings. For example, belief propagation works through
observed nodes, but there is no apparent way of generalising it for
unobserved nodes.
## **Theory independence**
More generally, given a GDAG $G$, we can look at the set of
distributions on $G$ that are generalised Markov with respect to a given
causal theory. Of particular importance are the following.
- The set $\mathcal {C}$ of generalised Markov distributions in
$Mat(\mathbb {R}_+)$ on $G$.
- The set $\mathcal {Q}$ of generalised Markov distributions in
$\mathsf{CPM}$ on $G$.
- The set $\mathcal {G}$ of all generalised Markov distributions on $G$.
(This is the set of generalised Markov distributions in
[Boxworld](https://www.nature.com/articles/nphys2916).)
Moreover, we can distinguish another class of distributions on $G$, by
not restricting to d-seperation of observed nodes, but considering
distributions that satisfy the observable conditional independences
given by any d-separation properties on the graph. Theorem 22 implies,
in particular that $G \subseteq I$.
And, so, since $Mat(\mathbb {R}_+)$ embeds into $\mathsf{CPM}$, we have
$\mathcal {C} \subseteq \mathcal {Q} \subseteq \mathcal {G} \subseteq \mathcal {I}$.
This means that one can ask for which graphs (some or all of) these
inequalities are strict, and the last part of the paper explores these
questions. In the [original paper](http://iopscience.iop.org/article/10.1088/1367-2630/16/11/113043/meta),
a sufficient condition is given for graphs to
satisfy $\mathcal {C} \neq \mathcal {I}$. I.e. for these graphs it is
guaranteed that the causal structure admits correlations that are
non-local. Moreover the authors show that their condition is necessary for
small enough graphs.
Another interesting result is that there exist graphs for which
$\mathcal {G} \neq \mathcal {I}$. This means that using a theory of
resources, whatever theory it may be, to explain correlations imposes
constraints that are stronger than those imposed by the relations
themselves.
## **What next?**
This setup represents one direction for using category theory to
generalise Bayesian networks. In our group work at the ACT workshop, we
considered another generalisation of Bayesian networks, this time
staying within the classical realm. Namely, building on the work of
[Bonchi, Gadducci, Kissinger, Sobocinski, and
Zanasi](https://arxiv.org/abs/1602.06771), we gave a functorial Markov
condition on directed graphs admitting cycles. Hopefully we'll present
this work here soon. In this case the Markov condition says that given that you know that
someone is having an allergic reaction, whether or not it is spring is
not going to influence your belief about the likelihood of them
sneezing. Which seems sensible.
Bayesian networks are useful
- as an inference tool, thanks to [belief
propagation](https://arxiv.org/abs/1201.4724) algorithms,
- and because, given a Bayesian network $(G,P)$, we can describe ['*d-separation*'](#d-separation) properties on $G$ which enable us to discover conditional
independences in $P$.
It is this second point that we'll be interested in here.
Before getting into the details of the paper, let's try to motivate this
discussion by explaining its title: *"Theory-independent limits on
correlations from generalized Bayesian networks\"* and giving a little
more background to the problem it aims to solve.
Crudely put, the paper aims to generalise a method that assumes
*classical mechanics* to one that holds in *quantum* and more general
theories.
Classical mechanics rests on two intuitively reasonable and desirable
assumptions, together called *local causality*,
- **Causality:**
Causality is usually treated as a physical primitive.
Simply put it is the principle that there is a (partial) ordering
of events in space time. In order to have information flow from
event $A$ to event $B$, $A$ *must* be in the past of $B$.
Physicists often define causality in terms of a *discarding*
principle: If we ignore the outcome of a physical process, it
doesn't matter what process has occurred. Or, put another way, the
outcome of a physical process doesn't change the initial conditions.
- **Locality:**
Locality is the assumption that, at any given instant, the values of
any particle's properties are independent of any other particle.
Intuitively, it says that particles are individual entities that can
be understood in isolation of any other particle.
Physicists usually picture particles as having a private list of
*numbers* determining their properties. The principle of locality
would be violated if any of the entries of such a list were *a
function* whose domain is another particle's property values.
In 1935 [Einstein, Podolsky and
Rosen](https://journals.aps.org/pr/pdf/10.1103/PhysRev.47.777) showed
that quantum mechanics (which was a recently born theory) predicted that
a pair of particles could be prepared so that applying an action on one
of them would instantaneously affect the other, no matter how distant in
space they were, thus contradicting local causality. This seemed so
unreasonable that the authors presented it as evidence that quantum
mechanics was wrong.
But Einstein was wrong. In 1964, [John S.
Bell](http://elib.bsu.by/bitstream/123456789/154530/1/1964-001%20P%20Bell%20-%20On%20the%20Einstein%20Podolsky%20Rosen%20paradox.pdf) set the bases for an
experimental test that would demonstrate that Einstein's "spooky action
at a distance" (Einstein's own words), now known as *entanglement*, was
indeed real. Bell's experiment has been replicated countless of times
and has plenty of variations. [This
video](https://www.youtube.com/watch?v=MUrXhAxGOWU) gives a detailed
explanation of one of these experiments, for a non-physicist audience.
But then, if acting on a particle has an instantaneous effect on a
distant point in space, one of the two principle above is violated: On
one hand, if we acted on both particles at the same time, each action
being a distinct event, both would be affecting each other's result, so
it would not be possible to decide on an ordering; causality would be
broken. The other option would be to reject locality: a property's value
may be given by *a function*, so the resulting value may
*instantaneously* change when the distant 'domain' particle is altered.
In that case, the particles' information was never separated in space,
as they were never truly isolated, so causality is preserved.
Since causality is integral to our understanding of the world and forms
the basis of scientific reasoning, the standard interpretation of
quantum mechanics is to accept non-locality.
The definition of Bayesian networks implies a [discarding
principle](#operational-probabilistic-theories) and hence there is a formal sense in which they are
causal (even if, as we shall see, the correlations they model do not
always reflect the temporal order). Under this interpretation, the
causal theory Bayesian networks describe is classical. Precisely, they
can only model probability distributions that satisfy local causality.
Hence, in particular, they are not sufficient to model all physical
correlations.
The goal of the paper is to develop a framework that generalises Bayesian
networks and d-separation results, so that we can still use graph
properties to reason about conditional dependence under any
given causal theory, be it classical, quantum, or even more general. In
particular, this theory will be able to handle all physically observed
correlations, and all theoretically postulated correlations.
Though category theory is not mentioned explicitly, the authors achieve
their goal by using the categorical framework of [*operational
probablistic theories* (OPTs)](https://arxiv.org/pdf/0908.1583.pdf).
## **Bayesian networks and d-separation**
Consider the situation in which we have three Boolean random variables.
Alice is either *sneezing* or she is not, she either has a a *fever* or
she does not, and she may or may not have *flu*.
Now, flu can cause both sneezing and fever, that is
$$P(sneezing \ | \ flu ) \neq P( sneezing) \ \text{ and likewise } \ P(fever \ | \ flu ) \neq P( fever)$$
so we could represent this graphically as
>
In this case the Markov condition says that given that you know that
someone is having an allergic reaction, whether or not it is spring is
not going to influence your belief about the likelihood of them
sneezing. Which seems sensible.
Bayesian networks are useful
- as an inference tool, thanks to [belief
propagation](https://arxiv.org/abs/1201.4724) algorithms,
- and because, given a Bayesian network $(G,P)$, we can describe ['*d-separation*'](#d-separation) properties on $G$ which enable us to discover conditional
independences in $P$.
It is this second point that we'll be interested in here.
Before getting into the details of the paper, let's try to motivate this
discussion by explaining its title: *"Theory-independent limits on
correlations from generalized Bayesian networks\"* and giving a little
more background to the problem it aims to solve.
Crudely put, the paper aims to generalise a method that assumes
*classical mechanics* to one that holds in *quantum* and more general
theories.
Classical mechanics rests on two intuitively reasonable and desirable
assumptions, together called *local causality*,
- **Causality:**
Causality is usually treated as a physical primitive.
Simply put it is the principle that there is a (partial) ordering
of events in space time. In order to have information flow from
event $A$ to event $B$, $A$ *must* be in the past of $B$.
Physicists often define causality in terms of a *discarding*
principle: If we ignore the outcome of a physical process, it
doesn't matter what process has occurred. Or, put another way, the
outcome of a physical process doesn't change the initial conditions.
- **Locality:**
Locality is the assumption that, at any given instant, the values of
any particle's properties are independent of any other particle.
Intuitively, it says that particles are individual entities that can
be understood in isolation of any other particle.
Physicists usually picture particles as having a private list of
*numbers* determining their properties. The principle of locality
would be violated if any of the entries of such a list were *a
function* whose domain is another particle's property values.
In 1935 [Einstein, Podolsky and
Rosen](https://journals.aps.org/pr/pdf/10.1103/PhysRev.47.777) showed
that quantum mechanics (which was a recently born theory) predicted that
a pair of particles could be prepared so that applying an action on one
of them would instantaneously affect the other, no matter how distant in
space they were, thus contradicting local causality. This seemed so
unreasonable that the authors presented it as evidence that quantum
mechanics was wrong.
But Einstein was wrong. In 1964, [John S.
Bell](http://elib.bsu.by/bitstream/123456789/154530/1/1964-001%20P%20Bell%20-%20On%20the%20Einstein%20Podolsky%20Rosen%20paradox.pdf) set the bases for an
experimental test that would demonstrate that Einstein's "spooky action
at a distance" (Einstein's own words), now known as *entanglement*, was
indeed real. Bell's experiment has been replicated countless of times
and has plenty of variations. [This
video](https://www.youtube.com/watch?v=MUrXhAxGOWU) gives a detailed
explanation of one of these experiments, for a non-physicist audience.
But then, if acting on a particle has an instantaneous effect on a
distant point in space, one of the two principle above is violated: On
one hand, if we acted on both particles at the same time, each action
being a distinct event, both would be affecting each other's result, so
it would not be possible to decide on an ordering; causality would be
broken. The other option would be to reject locality: a property's value
may be given by *a function*, so the resulting value may
*instantaneously* change when the distant 'domain' particle is altered.
In that case, the particles' information was never separated in space,
as they were never truly isolated, so causality is preserved.
Since causality is integral to our understanding of the world and forms
the basis of scientific reasoning, the standard interpretation of
quantum mechanics is to accept non-locality.
The definition of Bayesian networks implies a [discarding
principle](#operational-probabilistic-theories) and hence there is a formal sense in which they are
causal (even if, as we shall see, the correlations they model do not
always reflect the temporal order). Under this interpretation, the
causal theory Bayesian networks describe is classical. Precisely, they
can only model probability distributions that satisfy local causality.
Hence, in particular, they are not sufficient to model all physical
correlations.
The goal of the paper is to develop a framework that generalises Bayesian
networks and d-separation results, so that we can still use graph
properties to reason about conditional dependence under any
given causal theory, be it classical, quantum, or even more general. In
particular, this theory will be able to handle all physically observed
correlations, and all theoretically postulated correlations.
Though category theory is not mentioned explicitly, the authors achieve
their goal by using the categorical framework of [*operational
probablistic theories* (OPTs)](https://arxiv.org/pdf/0908.1583.pdf).
## **Bayesian networks and d-separation**
Consider the situation in which we have three Boolean random variables.
Alice is either *sneezing* or she is not, she either has a a *fever* or
she does not, and she may or may not have *flu*.
Now, flu can cause both sneezing and fever, that is
$$P(sneezing \ | \ flu ) \neq P( sneezing) \ \text{ and likewise } \ P(fever \ | \ flu ) \neq P( fever)$$
so we could represent this graphically as
>  Moreover, intuitively we wouldn't expect there to be any other edges in
the above graph. Sneezing and fever, though correlated - each is more
likely if Alice has flu - are not direct causes of each other. That is,
$$P(sneezing \ | \ fever ) \neq P(sneezing) \ \text{ but } \ P(sneezing \ | \ fever, \ flu ) = P(sneezing \ | \ flu).$$
### *Bayesian networks*
Let $G$ be a *directed acyclic graph* or *DAG* $G$. (Here a directed
graph is a [presheaf on
($\bullet \rightrightarrows \bullet$)](https://golem.ph.utexas.edu/category/2008/01/mark_weber_on_nerves_of_catego.html)).
The set $Pa(Y)$ of *parents* of a node $Y$ of $G$ contains those nodes
$X$ of $G$ such that there is a directed edge $X \to Y$.
So, in the example above $Pa(flu) = \emptyset$ while
$Pa(fever) = Pa(sneezing) = \{ flu \}$.
To each node $X$ of a directed graph $G$, we may associate a random
variable, also denoted $X$. If $V$ is the set of nodes of $G$ and
$(x_X)_{X \in V}$ is a choice of value $x_X$ for each node $X$, such
that $y$ is the chosen value for $Y$, then $pa(y)$ will denote the
$Pa(Y)$-tuple of values $(x_X)_{X \in Pa(Y)}$.
To define Bayesian networks, and establish the notation, let's revise
some probability basics.
Let $P(x,y \ | \ z)$ mean $P(X = x \text{ and } \ Y = y \ | \ Z = z)$,
the *probability that $X$ has the value $x$, and $Y$ has the value $y$
given that $Z$ has the value $z$*. Recall that this is given by
$$P(x,y \ |\ z) = \frac{ P(x,y,z) }{P(z)}.$$
The *chain rule* says that, given a value $x$ of $X$ and sets of values
$\Omega, \Lambda$ of other random variables,
$$P(x, \Omega \ | \ \Lambda) = P( x \ | \ \Lambda) P( \Omega \ | \ x, \Lambda).$$
Random variables $X$ and $Y$ are said to be *conditionally independent
given $Z$*, written $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$, if for all values
$x$ of $X$, $y$ of $Y$ and $z$ of $Z$
$$P(x,y \ | \ z) = P(x \ | \ z) P(y \ | \ z).$$
By the chain rule this is equivalent to
$$P(x \ | \ y,z ) = P (x \ | \ z) , \ \forall x,y, z.$$
More generally, we may replace $X,Y$ and $Z$ with sets of random
variables. So, in the special case that $Z$ is empty, then $X$ and $Y$
are independent if and only if $P(x, y) = P(x)P(y)$ for all $x,y$.
#### Markov condition
A joint probability distribution $P$ on the nodes of a DAG $G$ is said
to satisfy the Markov condition if for any set of random
variable $\{X_i\}_{i = 1}^n$ on the nodes of $G$, with choice of values
$\{x_i\}_{i = 1}^n$
$$P(x_i, \dots, x_n) = \prod_{i = 1}^n P(x_i \ | \ {pa(x_i)}).$$
So, for the flu, fever and sneezing example above, a distribution $P$
satisfies the Markov condition if
$$P(flu, \ fever, \ sneezing) = P(fever \ | \ flu) P(sneezing \ | \ flu) P(flu).$$
A Bayesian network is defined as a pair $(G,P)$ of a DAG $G$ and a joint
probability distribution $P$ on the nodes of $G$ that satisfies the
Markov condition with respect to $G$. This means that each node in a
Bayesian network is conditionally independent, given its parents, of any
of the remaining nodes.
In particular, given a Bayesian network $(G,P)$ such that there is a
directed edge $X \to Y$, the Markov condition implies that
$$\sum_{y} P(x,y) = \sum_y P(x) P(y \ | \ x) = P(x) \sum_y P(y \ | \ x) = P(x)$$
which may be interpreted as a [discard condition](#operational-probabilistic-theories). (The
ordering is reflected by the fact that we can't derive $P(y)$ from
$\sum_{x} P(x,y) = \sum_x P(x) P(y \ | \ x)$.)
Let's consider some simple examples.
**Fork**
In the example of flu, sneezing and fever above, the graph has a *fork*
shape. For a probability distribution $P$ to satisfy the Markov
condition for this graph we must have
$$P(x, y, z) = P(x \ | \ z) P(y \ | \ z)P(z), \ \forall x,y,z.$$
However, $P(x,y) \neq P(x) P(y)$.
In other words, $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$, though $X$ and $Y$ are
not independent. This makes sense, we wouldn't expect sneezing and fever
to be uncorrelated, but given that we know whether or not Alice has flu,
telling us that she has fever isn't going to tell us anything about her
sneezing.
**Collider**
Reversing the arrows in the fork graph above gives a *collider* as in
the following example.
>
Moreover, intuitively we wouldn't expect there to be any other edges in
the above graph. Sneezing and fever, though correlated - each is more
likely if Alice has flu - are not direct causes of each other. That is,
$$P(sneezing \ | \ fever ) \neq P(sneezing) \ \text{ but } \ P(sneezing \ | \ fever, \ flu ) = P(sneezing \ | \ flu).$$
### *Bayesian networks*
Let $G$ be a *directed acyclic graph* or *DAG* $G$. (Here a directed
graph is a [presheaf on
($\bullet \rightrightarrows \bullet$)](https://golem.ph.utexas.edu/category/2008/01/mark_weber_on_nerves_of_catego.html)).
The set $Pa(Y)$ of *parents* of a node $Y$ of $G$ contains those nodes
$X$ of $G$ such that there is a directed edge $X \to Y$.
So, in the example above $Pa(flu) = \emptyset$ while
$Pa(fever) = Pa(sneezing) = \{ flu \}$.
To each node $X$ of a directed graph $G$, we may associate a random
variable, also denoted $X$. If $V$ is the set of nodes of $G$ and
$(x_X)_{X \in V}$ is a choice of value $x_X$ for each node $X$, such
that $y$ is the chosen value for $Y$, then $pa(y)$ will denote the
$Pa(Y)$-tuple of values $(x_X)_{X \in Pa(Y)}$.
To define Bayesian networks, and establish the notation, let's revise
some probability basics.
Let $P(x,y \ | \ z)$ mean $P(X = x \text{ and } \ Y = y \ | \ Z = z)$,
the *probability that $X$ has the value $x$, and $Y$ has the value $y$
given that $Z$ has the value $z$*. Recall that this is given by
$$P(x,y \ |\ z) = \frac{ P(x,y,z) }{P(z)}.$$
The *chain rule* says that, given a value $x$ of $X$ and sets of values
$\Omega, \Lambda$ of other random variables,
$$P(x, \Omega \ | \ \Lambda) = P( x \ | \ \Lambda) P( \Omega \ | \ x, \Lambda).$$
Random variables $X$ and $Y$ are said to be *conditionally independent
given $Z$*, written $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$, if for all values
$x$ of $X$, $y$ of $Y$ and $z$ of $Z$
$$P(x,y \ | \ z) = P(x \ | \ z) P(y \ | \ z).$$
By the chain rule this is equivalent to
$$P(x \ | \ y,z ) = P (x \ | \ z) , \ \forall x,y, z.$$
More generally, we may replace $X,Y$ and $Z$ with sets of random
variables. So, in the special case that $Z$ is empty, then $X$ and $Y$
are independent if and only if $P(x, y) = P(x)P(y)$ for all $x,y$.
#### Markov condition
A joint probability distribution $P$ on the nodes of a DAG $G$ is said
to satisfy the Markov condition if for any set of random
variable $\{X_i\}_{i = 1}^n$ on the nodes of $G$, with choice of values
$\{x_i\}_{i = 1}^n$
$$P(x_i, \dots, x_n) = \prod_{i = 1}^n P(x_i \ | \ {pa(x_i)}).$$
So, for the flu, fever and sneezing example above, a distribution $P$
satisfies the Markov condition if
$$P(flu, \ fever, \ sneezing) = P(fever \ | \ flu) P(sneezing \ | \ flu) P(flu).$$
A Bayesian network is defined as a pair $(G,P)$ of a DAG $G$ and a joint
probability distribution $P$ on the nodes of $G$ that satisfies the
Markov condition with respect to $G$. This means that each node in a
Bayesian network is conditionally independent, given its parents, of any
of the remaining nodes.
In particular, given a Bayesian network $(G,P)$ such that there is a
directed edge $X \to Y$, the Markov condition implies that
$$\sum_{y} P(x,y) = \sum_y P(x) P(y \ | \ x) = P(x) \sum_y P(y \ | \ x) = P(x)$$
which may be interpreted as a [discard condition](#operational-probabilistic-theories). (The
ordering is reflected by the fact that we can't derive $P(y)$ from
$\sum_{x} P(x,y) = \sum_x P(x) P(y \ | \ x)$.)
Let's consider some simple examples.
**Fork**
In the example of flu, sneezing and fever above, the graph has a *fork*
shape. For a probability distribution $P$ to satisfy the Markov
condition for this graph we must have
$$P(x, y, z) = P(x \ | \ z) P(y \ | \ z)P(z), \ \forall x,y,z.$$
However, $P(x,y) \neq P(x) P(y)$.
In other words, $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$, though $X$ and $Y$ are
not independent. This makes sense, we wouldn't expect sneezing and fever
to be uncorrelated, but given that we know whether or not Alice has flu,
telling us that she has fever isn't going to tell us anything about her
sneezing.
**Collider**
Reversing the arrows in the fork graph above gives a *collider* as in
the following example.
>  Clearly whether or not Alice has allergies other than hayfever is
independent of what season it is. So we'd expect a distribution on this
graph to satisfy $X \perp\!\!\!\!\!\!\!\perp Y \ | \ \emptyset$. However, if
we know that Alice is having an allergic reaction, and it happens to be spring, we will likely
assume that she has some allergy, i.e. $X$ and $Y$ are not conditionally independent given $Z$.
Indeed, the Markov condition and chain rule for this graph gives us $X \perp\!\!\!\!\!\!\!\perp Y \ | \ \emptyset$:
$$P(x, y, z) = P(x)P(y) P(z \ | \ x,\ y) = P(z \ | \ x,\ y) P( x\ | \ y) P(y) \ \forall x,y,z.$$
from which we cannot derive
$P(x \ | \ z) P(y \ | \ z) = P(x,y \ | \ z)$. (However, it could still
be true for some particular choice of probability distribution.)
**Chain**
Finally, let us return to the *chain* of correlations presented in the
introduction.
Clearly the probabilities that it is spring and that Alice is sneezing
are not independent, and indeed, we cannot derive $P(x, y) = P(x) P(y)$.
However observe that, by the chain rule, a Markov distribution on the
chain graph must satisfy $X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$. If we know
Alice is having an allergic reaction that is not hayfever, whether or
not she is sneezing is not going to affect our guess as to what season
it is.
Crucially, in this case, knowing the season is also not going to affect
whether we think Alice is sneezing. By definition, conditional
independence of $X$ and $Y$ given $Z$ is symmetric in $X$ and $Y$. In
other words, a joint distribution $P$ on the variables $X,Y,Z$ satisfies
the Markov condition with respect to the chain graph
$$X \longrightarrow Z \longrightarrow Y$$
if and only if $P$ satisfies
the Markov condition on
$$Y \longrightarrow Z \longrightarrow X .$$
### *d-separation*
The above observations can be generalised to statements about
conditional independences in any Bayesian network. That is, if $(G,P)$
is a Bayesian network then the structure of $G$ is enough to derive all
the conditional independences in $P$ that are implied by the graph $G$
(in reality there may be more that have not been included in the
network!).
Given a DAG $G$ and a set of vertices $U$ of $G$, let $m(U)$ denote the
union of $U$ with all the vertices $v$ of $G$ such that there is a
directed edge from $U$ to $v$. The set $W(U)$ will denote the
*non-inclusive future* of $U$, that is, the set of vertices $v$ of $G$
for which there is no directed (possibly trivial) path from $v$ to $U$.
For a graph $G$, let $X, Y, Z$ now denote disjoint subsets of the
vertices of $G$ (and their corresponding random variables). Set
$W := W(X \cup Y \cup Z)$.
Then $X$ and $Y$ are said to be *d-separated* by $Z$, written
$X \perp Y \ | \ Z$, if there is a partition $\{U,V,W,Z\}$ of the nodes
of $G$ such that
- $X \subseteq U$ and $Y \subseteq V$, and
- $m(U) \cap m(V) \subseteq W,$ in other words $U$ and $V$ have no
direct influence on each other.
(This is lemma 19 in the paper.)
Now d-separation is really useful since it tells us everything there is
to know about the conditional dependences on Bayesian networks with
underlying graph $G$. Indeed,
#### Theorem 5
- **Soundness of d-separation** ([Verma and Pearl,
1988](https://www.sciencedirect.com/science/article/pii/B9780444886507500111))
If $P$ is a Markov distribution with respect to a graph $G$ then for
all disjoint subsets $X,Y,Z$ of nodes of $G$ $X \perp Y \ | \ Z$
implies that $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$.
- **Completeness of d-separation** ([Meek,
1995](https://dl.acm.org/citation.cfm?id=2074205)) If
$X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$ for all $P$ Markov with respect to
$G$, then $X \perp Y \ | \ Z$.
We can combine the previous examples of fork, collider and chain graphs
to get the following
>
Clearly whether or not Alice has allergies other than hayfever is
independent of what season it is. So we'd expect a distribution on this
graph to satisfy $X \perp\!\!\!\!\!\!\!\perp Y \ | \ \emptyset$. However, if
we know that Alice is having an allergic reaction, and it happens to be spring, we will likely
assume that she has some allergy, i.e. $X$ and $Y$ are not conditionally independent given $Z$.
Indeed, the Markov condition and chain rule for this graph gives us $X \perp\!\!\!\!\!\!\!\perp Y \ | \ \emptyset$:
$$P(x, y, z) = P(x)P(y) P(z \ | \ x,\ y) = P(z \ | \ x,\ y) P( x\ | \ y) P(y) \ \forall x,y,z.$$
from which we cannot derive
$P(x \ | \ z) P(y \ | \ z) = P(x,y \ | \ z)$. (However, it could still
be true for some particular choice of probability distribution.)
**Chain**
Finally, let us return to the *chain* of correlations presented in the
introduction.
Clearly the probabilities that it is spring and that Alice is sneezing
are not independent, and indeed, we cannot derive $P(x, y) = P(x) P(y)$.
However observe that, by the chain rule, a Markov distribution on the
chain graph must satisfy $X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$. If we know
Alice is having an allergic reaction that is not hayfever, whether or
not she is sneezing is not going to affect our guess as to what season
it is.
Crucially, in this case, knowing the season is also not going to affect
whether we think Alice is sneezing. By definition, conditional
independence of $X$ and $Y$ given $Z$ is symmetric in $X$ and $Y$. In
other words, a joint distribution $P$ on the variables $X,Y,Z$ satisfies
the Markov condition with respect to the chain graph
$$X \longrightarrow Z \longrightarrow Y$$
if and only if $P$ satisfies
the Markov condition on
$$Y \longrightarrow Z \longrightarrow X .$$
### *d-separation*
The above observations can be generalised to statements about
conditional independences in any Bayesian network. That is, if $(G,P)$
is a Bayesian network then the structure of $G$ is enough to derive all
the conditional independences in $P$ that are implied by the graph $G$
(in reality there may be more that have not been included in the
network!).
Given a DAG $G$ and a set of vertices $U$ of $G$, let $m(U)$ denote the
union of $U$ with all the vertices $v$ of $G$ such that there is a
directed edge from $U$ to $v$. The set $W(U)$ will denote the
*non-inclusive future* of $U$, that is, the set of vertices $v$ of $G$
for which there is no directed (possibly trivial) path from $v$ to $U$.
For a graph $G$, let $X, Y, Z$ now denote disjoint subsets of the
vertices of $G$ (and their corresponding random variables). Set
$W := W(X \cup Y \cup Z)$.
Then $X$ and $Y$ are said to be *d-separated* by $Z$, written
$X \perp Y \ | \ Z$, if there is a partition $\{U,V,W,Z\}$ of the nodes
of $G$ such that
- $X \subseteq U$ and $Y \subseteq V$, and
- $m(U) \cap m(V) \subseteq W,$ in other words $U$ and $V$ have no
direct influence on each other.
(This is lemma 19 in the paper.)
Now d-separation is really useful since it tells us everything there is
to know about the conditional dependences on Bayesian networks with
underlying graph $G$. Indeed,
#### Theorem 5
- **Soundness of d-separation** ([Verma and Pearl,
1988](https://www.sciencedirect.com/science/article/pii/B9780444886507500111))
If $P$ is a Markov distribution with respect to a graph $G$ then for
all disjoint subsets $X,Y,Z$ of nodes of $G$ $X \perp Y \ | \ Z$
implies that $X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$.
- **Completeness of d-separation** ([Meek,
1995](https://dl.acm.org/citation.cfm?id=2074205)) If
$X \perp\!\!\!\!\!\!\!\perp Y \ | \ Z$ for all $P$ Markov with respect to
$G$, then $X \perp Y \ | \ Z$.
We can combine the previous examples of fork, collider and chain graphs
to get the following
>  A priori, *Allergic reaction* is conditionally independent of *Fever*.
Indeed, we have the partition
>
A priori, *Allergic reaction* is conditionally independent of *Fever*.
Indeed, we have the partition
>  which clearly satisfies d-separation. However, if *Sneezing* is known then $W = \emptyset$,
so *Allergic reaction* and *Fever* are not independent. Indeed, if we use the same sets $U$ and $V$ as before, then
$m(U) \cap m(V) = \{ Sneezing \}$, so the condition for d-separation fails; and it does for any possible choice of $U$ and $V$.
Interestingly, if *Flu* is also known, we again obtain conditional independence between *Allergic reaction* and *Fever*, as shown below.
>
which clearly satisfies d-separation. However, if *Sneezing* is known then $W = \emptyset$,
so *Allergic reaction* and *Fever* are not independent. Indeed, if we use the same sets $U$ and $V$ as before, then
$m(U) \cap m(V) = \{ Sneezing \}$, so the condition for d-separation fails; and it does for any possible choice of $U$ and $V$.
Interestingly, if *Flu* is also known, we again obtain conditional independence between *Allergic reaction* and *Fever*, as shown below.
>  Before describing the limitations of this setup and why we may want to
generalise it, it is worth observing that Theorem 5 is genuinely useful
computationally. Theorem 5 says that given a Bayesian network $(G,P)$,
the structure of $G$ gives us a recipe to factor $P$, thereby greatly
increasing computation efficiency for Bayesian inference.
### *Latent variables, hidden variables, and unobservables*
In the context of Bayesian networks, there are two reasons that we may
wish to add variables to a probabilistic model, even if we are not
entirely sure what the variables signify or how they are distributed.
The first reason is statistical and the second is physical.
Consider the example of flu, fever and sneezing discussed earlier. Although our analysis told us $Fever \perp\!\!\!\!\!\!\!\perp Sneezing \ | \ Flu$, if we conduct an experiment we are likely to find:
$$P(fever \ | \ sneezing, \ flu) \neq P(fever \ | \ flu).$$
The problem is caused by the graph not properly modelling reality, but a simplification of it.
After all, there are a whole bunch of things that can cause sneezing and
flu. We just don't know what they all are or how to measure them. So, to
make the network work, we may add a hypothetical *latent variable* that
bunches together all the unknown joint causes, and equip it with a
distribution that makes the whole network Bayesian, so that we are still
able to perform inference methods like belief propagation.
>
Before describing the limitations of this setup and why we may want to
generalise it, it is worth observing that Theorem 5 is genuinely useful
computationally. Theorem 5 says that given a Bayesian network $(G,P)$,
the structure of $G$ gives us a recipe to factor $P$, thereby greatly
increasing computation efficiency for Bayesian inference.
### *Latent variables, hidden variables, and unobservables*
In the context of Bayesian networks, there are two reasons that we may
wish to add variables to a probabilistic model, even if we are not
entirely sure what the variables signify or how they are distributed.
The first reason is statistical and the second is physical.
Consider the example of flu, fever and sneezing discussed earlier. Although our analysis told us $Fever \perp\!\!\!\!\!\!\!\perp Sneezing \ | \ Flu$, if we conduct an experiment we are likely to find:
$$P(fever \ | \ sneezing, \ flu) \neq P(fever \ | \ flu).$$
The problem is caused by the graph not properly modelling reality, but a simplification of it.
After all, there are a whole bunch of things that can cause sneezing and
flu. We just don't know what they all are or how to measure them. So, to
make the network work, we may add a hypothetical *latent variable* that
bunches together all the unknown joint causes, and equip it with a
distribution that makes the whole network Bayesian, so that we are still
able to perform inference methods like belief propagation.
>  On the other hand, we may want to add variables to a Bayesian network if
we have evidence that doing so will provide a better model of reality.
For example, consider the network with just two connected nodes
>
On the other hand, we may want to add variables to a Bayesian network if
we have evidence that doing so will provide a better model of reality.
For example, consider the network with just two connected nodes
>  Every distribution on this graph is Markov, and we would expect there to
be a correlation between a road being wet and the grass next to it being
wet as well, but most people would claim that there's something missing
from the picture. After all, rain could be a 'common cause' of the road
and the grass being wet. So, it makes sense to add a third variable.
But maybe we can't observe whether it has rained or not, only whether
the grass and/or road are wet. Nonetheless, the correlation we observe
suggests that they have a common cause. To deal with such cases, we
could make the third variable *hidden*. We may not know what information
is included in a hidden variable, nor its probability distribution.
All that matters is that the hidden variable helps to explain the
observed correlations.
>
Every distribution on this graph is Markov, and we would expect there to
be a correlation between a road being wet and the grass next to it being
wet as well, but most people would claim that there's something missing
from the picture. After all, rain could be a 'common cause' of the road
and the grass being wet. So, it makes sense to add a third variable.
But maybe we can't observe whether it has rained or not, only whether
the grass and/or road are wet. Nonetheless, the correlation we observe
suggests that they have a common cause. To deal with such cases, we
could make the third variable *hidden*. We may not know what information
is included in a hidden variable, nor its probability distribution.
All that matters is that the hidden variable helps to explain the
observed correlations.
>  So, latent variables are a statistical tool that ensure the Markov
condition holds. Hence they are inherently classical, and can, in
theory, be known. But the universe is not classical, so, even if we lump
whatever we want into as many classical hidden variables as we want and
put them wherever we need, in some cases, there will still be
empirically observed correlations that do not satisfy the Markov
condition.
Most famously, Bell's experiment shows that it is possible to have
distinct variables $A$ and $B$ that exhibit correlations that cannot be
explained by any classical hidden variable, since classical variables
are restricted by the principle of locality.
In other words, though $A \perp B \ | \ \Lambda$,
$$P(a \ | b,\ \lambda) \neq P(a \ | \ \lambda).$$
Implicitly, this means that a *classical* $\Lambda$ is not enough. If we
want $P(a \ | b,\ \lambda) \neq P(a \ | \ \lambda)$ to hold, $\Lambda$
must be a non-local (non-classical) variable. Quantum mechanics implies
that we can't possibly empirically find the value of a non-local
variable (for similar reasons to the Heisenberg's uncertainty
principle), so non-classical variables are often called *unobservables*.
In particular, it is irrelevant to question whether
$A \perp\!\!\!\!\!\!\!\perp B \ | \ \Lambda$, as we would need to know the value
of $\Lambda$ in order to condition over it.
Indeed, this is the key idea behind what follows. We declare certain
variables to be unobservable and then insist that conditional (in)dependence
only makes sense between *observable variables conditioned
over observable variables*.
## **Generalising classical causality**
The correlations observed in the Bell experiment can be explained by
quantum mechanics. But thought experiments such as the one described
[here](https://www.nature.com/articles/nphys2916) suggest that
theoretically, correlations may exist that violate even quantum
causality.
So, given that graphical models and d-separation provide such a powerful
tool for causal reasoning in the classical context, how can we
generalise the Markov condition and Theorem 5 to quantum, and even more
general causal theories? And, if we have a *theory-independent* Markov
condition, are there d-separation results that don't correspond to any
given causal theory?
Clearly the first step in answering these questions is to fix a
definition of a *causal* theory.
### *Operational probabalistic theories*
An [*operational theory*](https://arxiv.org/pdf/0908.1583.pdf) is a
symmetric monoidal category $(\mathsf {C}, \otimes, I)$ whose objects are
known as *systems* or *resources*. Morphisms are finite sets
$f = \{\mathcal {C}_i\}_{i \in I}$ called *tests*, whose elements are
called *outcomes*. Tests with a single element are called
*deterministic*, and for each system $A \in ob (\mathsf {C})$, the
identity $id_A \in \mathsf (A,A)$ is a deterministic test.
In this discussion, we'll identify tests
$\{\mathcal {C}_i \}_i , \{\mathcal {D}_j\}_j$ in $\mathsf {C}$ if we may
always replace one with the other without affecting the distributions in
$\mathsf {C}(I, I)$.
Given $\{\mathcal {C}_i \}_i \in \mathsf {C}(B, C)$ and
$\{\mathcal {D}_j \} \in \mathsf {C}(A, B)$, their composition $f \circ g$
is given by
$$\{ \mathcal {C}_i \circ \mathcal {D}_j \}_{i,j} \in \mathsf {C}(A, C).$$
First apply $\mathcal {D}$ with output $B$ then apply $\mathcal {C}$ with
outcome $C$.
The monoidal composition
$\{ \mathcal {C}_i \otimes \mathcal {D}_j \}_{i, j} \in \mathsf {C}(A \otimes C, B \otimes D)$
corresponds to applying $\{\mathcal {C}_i\}_i \in \mathsf {C}(A,B)$ and
$\{ \mathcal {D}_j \}_j$ separately on $A$ and $C$.
An *operational probabilistic theory* or *OPT* is an operational theory
such that every test $I \to I$ is a probability distribution.
A morphism $\{ \mathcal {C}_i \}_i \in \mathsf {C}(A, I)$ is called an
*effect* on $A$. An OPT $\mathsf {C}$ is called *causal* or a *causal
theory* if, for each system $A \in ob (\mathsf {C})$, there is a unique
deterministic effect $\top_A \in \mathsf {C}( A, I)$ which we call the
*discard* of $A$.
In particular, for a causal OPT $\mathsf {C}$, uniqueness of the discard
implies that, for all systems $A, B \in ob (\mathsf {C})$,
$$\top_A \otimes \top_B = \top_{A \otimes B},$$
and, given any
determinstic test $\mathcal {C} \in \mathsf {C}(A, B)$,
$$\top_B \circ \mathcal {C} = \top_A.$$
The existence of a discard map allows a definition of *causal morphisms*
in a causal theory. For example, as we saw in
[January](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
when we discussed [Kissinger and Uijlen's
paper](https://arxiv.org/abs/1701.04732), a test
$\{ \mathcal {C}_i \}_i \in \mathsf {C} (A, B)$ is *causal* if
$$\top_B \circ \{ \mathcal {C}_i \}_i = \top_A \in \mathsf {C}( A, I).$$
In other words, for a causal test, discarding the outcome is the same as
not performing the test. Intuitively it is not obvious why such
morphisms should be called causal. But this definition enables the
formulation of a [*non-signalling
condition*](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
that describes the conditions under which the possibility of
cause-effect correlation is excluded, in particular, it implies the
impossibility of time travel.
#### Examples
The category
[$Mat(\mathbb {R}_+)$](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
of natural numbers and with $Mat(\mathbb {R}_+)(m,n)$ the set of
$n \times m$ matrices, has the structure of a causal OPT. The causal
morphisms in $Mat(\mathbb {R}_+)$ are the stochastic maps (the matrices
whose columns sum to 1). This category describes classical probability
theory.
The category [$\mathsf{CPM}$](https://arxiv.org/abs/1701.04732) of sets
of linear operators on Hilbert spaces and completely positive maps
between them is an OPT and describes quantum relations. The causal
morphisms are the trace preserving completely positive maps.
Finally, [Boxworld](https://www.nature.com/articles/nphys2916) is the
theory that allows to describe any correlation between two
variables as the cause of some resource of the theory in the past.
### *Generalised Bayesian networks*
So, we're finally ready to give the main construction and results of the
paper. As mentioned before, to get a generalised d-separation result,
the idea is that we will distinguish observable and unobservable
variables, and simply insist that conditional independence is only
defined relative to observable variables.
To this end, a *generalised DAG* or *GDAG* is a DAG $G$ together with a
partition on the nodes of $G$ into two subsets called *observed* and
*unobserved*. We'll represent observed nodes by triangles, and
unobserved nodes by circles. An edge out of an (un)observed node will be
called *(un)observed* and represented by a (solid) dashed arrow.
In order to get a generalisation of Theorem 5, we still need to come up
with a sensible generalisation of the Markov property which will
essentially say that at an observed node that has only observed parents,
the distribution must be Markov. However, if an observed node has an
unobserved parent, the latter's whole history is needed to describe the
distribution.
To state this precisely, we will associate a causal theory
$(\mathsf {C}, \otimes, I)$ to a GDAG $G$ via an assignment of systems to
edges of $G$ and tests to nodes of $G$, such that the observed edges of
$G$ will 'carry' only the outcomes of classical tests (so will say
something about conditional probability) whereas unobserved edges will
carry only the output system.
Precisely, such an assignment $P$ satisfies the *generalised Markov
condition (GMC)* and is called a *generalised Markov distribution* if
- Each unobserved edge corresponds to a distinct system in the theory.
- *If we can't observe what is happening at a node, we can't condition
over it:* To each unobserved node and each value of its observed
parents, we assign a deterministic test from the system defined by
the product of its incoming (unobserved) edges to the system defined
by the product of its outgoing (unobserved) edges.
- Each observed node $X$ is an observation test, i.e. a morphism in
$\mathsf {C}(A, I)$ for the system $A \in ob( \mathsf {C})$
corresponding to the product of the systems assigned to the
unobserved input edges of $X$. Since $\mathsf {C}$ is a causal theory,
this says that $X$ is assigned a classical random variable, also
denoted $X$, and that if $Y$ is an observed node, and has observed
parent $X$, the distribution at $Y$ is conditionally dependent on
the distribution at $X$ (see
[here](https://arxiv.org/pdf/0908.1583.pdf) for details).
- It therefore follows that each observed edge is assigned the trivial system $I$.
- The joint probability distribution on the observed nodes of $G$ is given by the morphism $\mathsf {C}(I, I)$
that results from these assignments.
A *generalised Bayesian network* consists of a GDAG $G$ together with a
generalised Markov distribution $P$ on $G$.
#### Example
Consider the following GDAG
>
So, latent variables are a statistical tool that ensure the Markov
condition holds. Hence they are inherently classical, and can, in
theory, be known. But the universe is not classical, so, even if we lump
whatever we want into as many classical hidden variables as we want and
put them wherever we need, in some cases, there will still be
empirically observed correlations that do not satisfy the Markov
condition.
Most famously, Bell's experiment shows that it is possible to have
distinct variables $A$ and $B$ that exhibit correlations that cannot be
explained by any classical hidden variable, since classical variables
are restricted by the principle of locality.
In other words, though $A \perp B \ | \ \Lambda$,
$$P(a \ | b,\ \lambda) \neq P(a \ | \ \lambda).$$
Implicitly, this means that a *classical* $\Lambda$ is not enough. If we
want $P(a \ | b,\ \lambda) \neq P(a \ | \ \lambda)$ to hold, $\Lambda$
must be a non-local (non-classical) variable. Quantum mechanics implies
that we can't possibly empirically find the value of a non-local
variable (for similar reasons to the Heisenberg's uncertainty
principle), so non-classical variables are often called *unobservables*.
In particular, it is irrelevant to question whether
$A \perp\!\!\!\!\!\!\!\perp B \ | \ \Lambda$, as we would need to know the value
of $\Lambda$ in order to condition over it.
Indeed, this is the key idea behind what follows. We declare certain
variables to be unobservable and then insist that conditional (in)dependence
only makes sense between *observable variables conditioned
over observable variables*.
## **Generalising classical causality**
The correlations observed in the Bell experiment can be explained by
quantum mechanics. But thought experiments such as the one described
[here](https://www.nature.com/articles/nphys2916) suggest that
theoretically, correlations may exist that violate even quantum
causality.
So, given that graphical models and d-separation provide such a powerful
tool for causal reasoning in the classical context, how can we
generalise the Markov condition and Theorem 5 to quantum, and even more
general causal theories? And, if we have a *theory-independent* Markov
condition, are there d-separation results that don't correspond to any
given causal theory?
Clearly the first step in answering these questions is to fix a
definition of a *causal* theory.
### *Operational probabalistic theories*
An [*operational theory*](https://arxiv.org/pdf/0908.1583.pdf) is a
symmetric monoidal category $(\mathsf {C}, \otimes, I)$ whose objects are
known as *systems* or *resources*. Morphisms are finite sets
$f = \{\mathcal {C}_i\}_{i \in I}$ called *tests*, whose elements are
called *outcomes*. Tests with a single element are called
*deterministic*, and for each system $A \in ob (\mathsf {C})$, the
identity $id_A \in \mathsf (A,A)$ is a deterministic test.
In this discussion, we'll identify tests
$\{\mathcal {C}_i \}_i , \{\mathcal {D}_j\}_j$ in $\mathsf {C}$ if we may
always replace one with the other without affecting the distributions in
$\mathsf {C}(I, I)$.
Given $\{\mathcal {C}_i \}_i \in \mathsf {C}(B, C)$ and
$\{\mathcal {D}_j \} \in \mathsf {C}(A, B)$, their composition $f \circ g$
is given by
$$\{ \mathcal {C}_i \circ \mathcal {D}_j \}_{i,j} \in \mathsf {C}(A, C).$$
First apply $\mathcal {D}$ with output $B$ then apply $\mathcal {C}$ with
outcome $C$.
The monoidal composition
$\{ \mathcal {C}_i \otimes \mathcal {D}_j \}_{i, j} \in \mathsf {C}(A \otimes C, B \otimes D)$
corresponds to applying $\{\mathcal {C}_i\}_i \in \mathsf {C}(A,B)$ and
$\{ \mathcal {D}_j \}_j$ separately on $A$ and $C$.
An *operational probabilistic theory* or *OPT* is an operational theory
such that every test $I \to I$ is a probability distribution.
A morphism $\{ \mathcal {C}_i \}_i \in \mathsf {C}(A, I)$ is called an
*effect* on $A$. An OPT $\mathsf {C}$ is called *causal* or a *causal
theory* if, for each system $A \in ob (\mathsf {C})$, there is a unique
deterministic effect $\top_A \in \mathsf {C}( A, I)$ which we call the
*discard* of $A$.
In particular, for a causal OPT $\mathsf {C}$, uniqueness of the discard
implies that, for all systems $A, B \in ob (\mathsf {C})$,
$$\top_A \otimes \top_B = \top_{A \otimes B},$$
and, given any
determinstic test $\mathcal {C} \in \mathsf {C}(A, B)$,
$$\top_B \circ \mathcal {C} = \top_A.$$
The existence of a discard map allows a definition of *causal morphisms*
in a causal theory. For example, as we saw in
[January](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
when we discussed [Kissinger and Uijlen's
paper](https://arxiv.org/abs/1701.04732), a test
$\{ \mathcal {C}_i \}_i \in \mathsf {C} (A, B)$ is *causal* if
$$\top_B \circ \{ \mathcal {C}_i \}_i = \top_A \in \mathsf {C}( A, I).$$
In other words, for a causal test, discarding the outcome is the same as
not performing the test. Intuitively it is not obvious why such
morphisms should be called causal. But this definition enables the
formulation of a [*non-signalling
condition*](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
that describes the conditions under which the possibility of
cause-effect correlation is excluded, in particular, it implies the
impossibility of time travel.
#### Examples
The category
[$Mat(\mathbb {R}_+)$](https://golem.ph.utexas.edu/category/2018/01/a_categorical_semantics_for_ca.html#more)
of natural numbers and with $Mat(\mathbb {R}_+)(m,n)$ the set of
$n \times m$ matrices, has the structure of a causal OPT. The causal
morphisms in $Mat(\mathbb {R}_+)$ are the stochastic maps (the matrices
whose columns sum to 1). This category describes classical probability
theory.
The category [$\mathsf{CPM}$](https://arxiv.org/abs/1701.04732) of sets
of linear operators on Hilbert spaces and completely positive maps
between them is an OPT and describes quantum relations. The causal
morphisms are the trace preserving completely positive maps.
Finally, [Boxworld](https://www.nature.com/articles/nphys2916) is the
theory that allows to describe any correlation between two
variables as the cause of some resource of the theory in the past.
### *Generalised Bayesian networks*
So, we're finally ready to give the main construction and results of the
paper. As mentioned before, to get a generalised d-separation result,
the idea is that we will distinguish observable and unobservable
variables, and simply insist that conditional independence is only
defined relative to observable variables.
To this end, a *generalised DAG* or *GDAG* is a DAG $G$ together with a
partition on the nodes of $G$ into two subsets called *observed* and
*unobserved*. We'll represent observed nodes by triangles, and
unobserved nodes by circles. An edge out of an (un)observed node will be
called *(un)observed* and represented by a (solid) dashed arrow.
In order to get a generalisation of Theorem 5, we still need to come up
with a sensible generalisation of the Markov property which will
essentially say that at an observed node that has only observed parents,
the distribution must be Markov. However, if an observed node has an
unobserved parent, the latter's whole history is needed to describe the
distribution.
To state this precisely, we will associate a causal theory
$(\mathsf {C}, \otimes, I)$ to a GDAG $G$ via an assignment of systems to
edges of $G$ and tests to nodes of $G$, such that the observed edges of
$G$ will 'carry' only the outcomes of classical tests (so will say
something about conditional probability) whereas unobserved edges will
carry only the output system.
Precisely, such an assignment $P$ satisfies the *generalised Markov
condition (GMC)* and is called a *generalised Markov distribution* if
- Each unobserved edge corresponds to a distinct system in the theory.
- *If we can't observe what is happening at a node, we can't condition
over it:* To each unobserved node and each value of its observed
parents, we assign a deterministic test from the system defined by
the product of its incoming (unobserved) edges to the system defined
by the product of its outgoing (unobserved) edges.
- Each observed node $X$ is an observation test, i.e. a morphism in
$\mathsf {C}(A, I)$ for the system $A \in ob( \mathsf {C})$
corresponding to the product of the systems assigned to the
unobserved input edges of $X$. Since $\mathsf {C}$ is a causal theory,
this says that $X$ is assigned a classical random variable, also
denoted $X$, and that if $Y$ is an observed node, and has observed
parent $X$, the distribution at $Y$ is conditionally dependent on
the distribution at $X$ (see
[here](https://arxiv.org/pdf/0908.1583.pdf) for details).
- It therefore follows that each observed edge is assigned the trivial system $I$.
- The joint probability distribution on the observed nodes of $G$ is given by the morphism $\mathsf {C}(I, I)$
that results from these assignments.
A *generalised Bayesian network* consists of a GDAG $G$ together with a
generalised Markov distribution $P$ on $G$.
#### Example
Consider the following GDAG
>  Let's build its OPT morphism as indictated by the *generalised Markov condition*.
The observed node $X$ has no incoming edges so it corresponds to a $\mathsf {C}(I, I)$ morphism, and thus we assign a probability
distribution to it.
The unobserved node A *depends* on $X$, and has no unobserved inputs, so
we assign a deterministic test $A(x): I \to A$ for each value $x$ of
$X$.
>
Let's build its OPT morphism as indictated by the *generalised Markov condition*.
The observed node $X$ has no incoming edges so it corresponds to a $\mathsf {C}(I, I)$ morphism, and thus we assign a probability
distribution to it.
The unobserved node A *depends* on $X$, and has no unobserved inputs, so
we assign a deterministic test $A(x): I \to A$ for each value $x$ of
$X$.
> 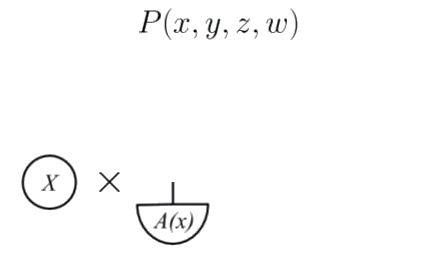 The observed node $Y$ has one incoming unobserved edge and no incoming
observed edges so we assign to it a test $Y: A \to I$ such that, for
each value $x$ of $X$, $Y \circ A(x)$ is a probability distribution.
Building up the rest of the picture gives an OPT diagram of the form
>
The observed node $Y$ has one incoming unobserved edge and no incoming
observed edges so we assign to it a test $Y: A \to I$ such that, for
each value $x$ of $X$, $Y \circ A(x)$ is a probability distribution.
Building up the rest of the picture gives an OPT diagram of the form
>  which is a $\mathsf {C}(I, I)$ morphism that defines the joint probability distribution $P(x,y,z,w)$.
We now have all the ingredients to state Theorem 22, the *generalised
d-separation theorem*. This is the analogue of Theorem 5 for generalised
Markov distributions.
#### Theorem 22
Given a GDAG $G$ and subsets $X,Y, Z$ of observed nodes
- if a probability distribution $P$ is generalised Markov relative to
$G$ then
$X \perp Y \ | \ Z \Rightarrow X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$.
- If $X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$ holds for all generalised Markov
probability distributions on $G$, then $X \perp Y \ | \ Z$.
Note in particular that there is no change in the definition of
d-separation: d-separation of a GDAG $G$ is simply d-separation with
respect to its underlying DAG. There is also no change in the definition
of conditional independence. Now, however, we restrict to statements of
conditional independence with respect to observed nodes only. This
enables the generalised soundness and completeness statements of the
theorem.
The proof of soundness uses uniqueness of discarding, and completeness
follows since generalised Markov is a stronger condition on a
distribution than classically Markov.
### *Classical distributions on GDAGs*
Theorem 22 is all well and good. But does it really generalise the
classical case? That is, can we recover Theorem 5 for all classical
Bayesian networks from Theorem 22?
As a first step, Proposition 17 states that if all the nodes of a
generalised Bayesian network are observed, then it is a classical
bayesian network. In fact, this follows pretty immediately from the
definitions.
Moreover, it is easily checked that, given a classical Bayesian network,
even if it has hidden or latent variables, it can still be expressed
directly as a generalised Bayesian network with no unobserved nodes.
In fact, Theorem 22 generalises Theorem 5 in a stricter sense. That is,
the generalised Bayesian network setup together with classical causality
adds nothing extra to the theory of classical Bayesian networks. If a
generalised Markov distribution is classical (then hidden and latent
variables may be represented by unobserved nodes), it can be viewed as a
classical Bayesian network. More precisely, Lemma 18 says that, given
any generalised Bayesian network $(G,P)$ with underlying DAG $G'$ and
distribution $P \in \mathcal {C}$, we can construct a classical Bayesian
network $(G', P')$ such that $P'$ agrees with $P$ on the observed nodes.
It is worth voicing a note of caution. The authors themselves
mention in the conclusion that the construction based on GDAGs with two
types of nodes is not entirely satisfactory. The problem is that,
although the setups and results presented here do give a generalisation
of Theorem 5, they do not, as such, provide a way of generalising
Bayesian networks as they are used for probabalistic inference to
non-classical settings. For example, belief propagation works through
observed nodes, but there is no apparent way of generalising it for
unobserved nodes.
## **Theory independence**
More generally, given a GDAG $G$, we can look at the set of
distributions on $G$ that are generalised Markov with respect to a given
causal theory. Of particular importance are the following.
- The set $\mathcal {C}$ of generalised Markov distributions in
$Mat(\mathbb {R}_+)$ on $G$.
- The set $\mathcal {Q}$ of generalised Markov distributions in
$\mathsf{CPM}$ on $G$.
- The set $\mathcal {G}$ of all generalised Markov distributions on $G$.
(This is the set of generalised Markov distributions in
[Boxworld](https://www.nature.com/articles/nphys2916).)
Moreover, we can distinguish another class of distributions on $G$, by
not restricting to d-seperation of observed nodes, but considering
distributions that satisfy the observable conditional independences
given by any d-separation properties on the graph. Theorem 22 implies,
in particular that $G \subseteq I$.
And, so, since $Mat(\mathbb {R}_+)$ embeds into $\mathsf{CPM}$, we have
$\mathcal {C} \subseteq \mathcal {Q} \subseteq \mathcal {G} \subseteq \mathcal {I}$.
This means that one can ask for which graphs (some or all of) these
inequalities are strict, and the last part of the paper explores these
questions. In the [original paper](http://iopscience.iop.org/article/10.1088/1367-2630/16/11/113043/meta),
a sufficient condition is given for graphs to
satisfy $\mathcal {C} \neq \mathcal {I}$. I.e. for these graphs it is
guaranteed that the causal structure admits correlations that are
non-local. Moreover the authors show that their condition is necessary for
small enough graphs.
Another interesting result is that there exist graphs for which
$\mathcal {G} \neq \mathcal {I}$. This means that using a theory of
resources, whatever theory it may be, to explain correlations imposes
constraints that are stronger than those imposed by the relations
themselves.
## **What next?**
This setup represents one direction for using category theory to
generalise Bayesian networks. In our group work at the ACT workshop, we
considered another generalisation of Bayesian networks, this time
staying within the classical realm. Namely, building on the work of
[Bonchi, Gadducci, Kissinger, Sobocinski, and
Zanasi](https://arxiv.org/abs/1602.06771), we gave a functorial Markov
condition on directed graphs admitting cycles. Hopefully we'll present
this work here soon.
which is a $\mathsf {C}(I, I)$ morphism that defines the joint probability distribution $P(x,y,z,w)$.
We now have all the ingredients to state Theorem 22, the *generalised
d-separation theorem*. This is the analogue of Theorem 5 for generalised
Markov distributions.
#### Theorem 22
Given a GDAG $G$ and subsets $X,Y, Z$ of observed nodes
- if a probability distribution $P$ is generalised Markov relative to
$G$ then
$X \perp Y \ | \ Z \Rightarrow X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$.
- If $X\perp\!\!\!\!\!\!\!\perp Y \ | \ Z$ holds for all generalised Markov
probability distributions on $G$, then $X \perp Y \ | \ Z$.
Note in particular that there is no change in the definition of
d-separation: d-separation of a GDAG $G$ is simply d-separation with
respect to its underlying DAG. There is also no change in the definition
of conditional independence. Now, however, we restrict to statements of
conditional independence with respect to observed nodes only. This
enables the generalised soundness and completeness statements of the
theorem.
The proof of soundness uses uniqueness of discarding, and completeness
follows since generalised Markov is a stronger condition on a
distribution than classically Markov.
### *Classical distributions on GDAGs*
Theorem 22 is all well and good. But does it really generalise the
classical case? That is, can we recover Theorem 5 for all classical
Bayesian networks from Theorem 22?
As a first step, Proposition 17 states that if all the nodes of a
generalised Bayesian network are observed, then it is a classical
bayesian network. In fact, this follows pretty immediately from the
definitions.
Moreover, it is easily checked that, given a classical Bayesian network,
even if it has hidden or latent variables, it can still be expressed
directly as a generalised Bayesian network with no unobserved nodes.
In fact, Theorem 22 generalises Theorem 5 in a stricter sense. That is,
the generalised Bayesian network setup together with classical causality
adds nothing extra to the theory of classical Bayesian networks. If a
generalised Markov distribution is classical (then hidden and latent
variables may be represented by unobserved nodes), it can be viewed as a
classical Bayesian network. More precisely, Lemma 18 says that, given
any generalised Bayesian network $(G,P)$ with underlying DAG $G'$ and
distribution $P \in \mathcal {C}$, we can construct a classical Bayesian
network $(G', P')$ such that $P'$ agrees with $P$ on the observed nodes.
It is worth voicing a note of caution. The authors themselves
mention in the conclusion that the construction based on GDAGs with two
types of nodes is not entirely satisfactory. The problem is that,
although the setups and results presented here do give a generalisation
of Theorem 5, they do not, as such, provide a way of generalising
Bayesian networks as they are used for probabalistic inference to
non-classical settings. For example, belief propagation works through
observed nodes, but there is no apparent way of generalising it for
unobserved nodes.
## **Theory independence**
More generally, given a GDAG $G$, we can look at the set of
distributions on $G$ that are generalised Markov with respect to a given
causal theory. Of particular importance are the following.
- The set $\mathcal {C}$ of generalised Markov distributions in
$Mat(\mathbb {R}_+)$ on $G$.
- The set $\mathcal {Q}$ of generalised Markov distributions in
$\mathsf{CPM}$ on $G$.
- The set $\mathcal {G}$ of all generalised Markov distributions on $G$.
(This is the set of generalised Markov distributions in
[Boxworld](https://www.nature.com/articles/nphys2916).)
Moreover, we can distinguish another class of distributions on $G$, by
not restricting to d-seperation of observed nodes, but considering
distributions that satisfy the observable conditional independences
given by any d-separation properties on the graph. Theorem 22 implies,
in particular that $G \subseteq I$.
And, so, since $Mat(\mathbb {R}_+)$ embeds into $\mathsf{CPM}$, we have
$\mathcal {C} \subseteq \mathcal {Q} \subseteq \mathcal {G} \subseteq \mathcal {I}$.
This means that one can ask for which graphs (some or all of) these
inequalities are strict, and the last part of the paper explores these
questions. In the [original paper](http://iopscience.iop.org/article/10.1088/1367-2630/16/11/113043/meta),
a sufficient condition is given for graphs to
satisfy $\mathcal {C} \neq \mathcal {I}$. I.e. for these graphs it is
guaranteed that the causal structure admits correlations that are
non-local. Moreover the authors show that their condition is necessary for
small enough graphs.
Another interesting result is that there exist graphs for which
$\mathcal {G} \neq \mathcal {I}$. This means that using a theory of
resources, whatever theory it may be, to explain correlations imposes
constraints that are stronger than those imposed by the relations
themselves.
## **What next?**
This setup represents one direction for using category theory to
generalise Bayesian networks. In our group work at the ACT workshop, we
considered another generalisation of Bayesian networks, this time
staying within the classical realm. Namely, building on the work of
[Bonchi, Gadducci, Kissinger, Sobocinski, and
Zanasi](https://arxiv.org/abs/1602.06771), we gave a functorial Markov
condition on directed graphs admitting cycles. Hopefully we'll present
this work here soon.